1. Background
Enterococci can be considered as gastrointestinal and vaginal flora (1). However, these bacteria are also considered as the third most common cause of nosocomial infections, which are now spreading and are hard to treat around the world included Iran (2). Enterococci can invade bloodstream and can cause urinary tract infection, endocarditis, peritonitis, and many other types of infections (2, 3). Antimicrobial resistance of these organisms has been a concerning issue for a long time (1-4). These organisms are inherently resistant to cephalosporins, clindamycin, and many other antibiotics. Beta-lactamase production by some strains of Enterococcusfaecalis and Enterococcusfaecium is also another mechanism of resistance. Vancomycin-resistant Enterococcus (VRE) not only show high-grade resistance to vancomycin and aminoglycosides, but also may show resistance to penicillins (3).
Resistance to vancomycin in these organisms is an important problem because this antibiotic is very effective against gram-positive bacteria whose rate of resistance has been increasing during recent years due to increase in the presence of plasmid bearing resistance genes (5). The resistance inducing gene for vancomycin is called van gene and is subtyped to type A and B on transposon Tn1546. These genes can be potentially introduced to conjugative plasmid, transferred within enterococcal strains as well as to the other organisms such as staphylococci, and can increase the potential risk of vancomycin-resistant Staphylococcus (VRS) in the community. These two genes cause high-grade resistance to vancomycin while genes type D, C, and E cause low-grade resistance to vancomycin and are located on chromosome (6, 7). Virulence factors specific to glycopeptide-resistant Enterococcus have not been identified so far (5). It seems that their virulence is similar to vancomycin-sensitive strains and although the mortality rate is higher, it can be attributed to failure of on time treatment. Strains positive for vanA are usually resistant to both vancomycin and teicoplanin but vanB positive strains usually respond to the later one (5). Therefore, determination of these resistance genes is important for therapeutic strategies.
2. Objectives
In this study, we looked for resistance genes of vanA and vanB in VRE isolated from intestinal colonization of children admitted to the pediatric and neonates intensive care units (PICU and NICU, respectively) of Ali-Asghar Children’s Hospital during 2012-2013. The Ethical Committee of Pediatric Infections Research Center of Mofid Children’s Hospital approved the study protocol.
3. Patients and Methods
3.1. Bacterial Identification and Patients Selection
This descriptive study was conducted from January 2012 to June 2013. Surveillance of VRE colonization (rectal or stool swab) was performed on all children aged 18 months or younger admitted to the PICU and NICU of Ali-Asghar Children's Hospital, Tehran, who met the inclusion criteria. The inclusion criteria were serious systemic illness including admission to NICU or PICU for at least one week, malignancy, chronic kidney, lung, or liver diseases, treatment with chemotherapeutic agents, immunodeficiency, treatment with high-dose corticosteroids (more than 1 mg/kg/day for more than one month), malnutrition (body weight < 5th percentile for age), and previous treatment with second or third generation cephalosporins, aminoglycosides, or broad-spectrum β-lactams within the past three months. We exclude children with proven enterococcal infection and children whose parents were not willing to participate in this study.
Rectal swab samples were transferred to the Pediatric Infections Research Center, Mofid Children’s Hospital, immediately after obtaining. Enterococci were diagnosed in samples by Gram staining, biochemical tests like catalase, growing on bile esculin agar and NaCl (6.5%) media, and ability to growth on selective media of Enterococcosel agar.
3.2. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility to ampicillin (10 μg), penicillin (10 μg), ciprofloxacin (5 μg), rifampin (5 μg), teicoplanin (30 μg), chloramphenicol (30 μg), vancomycin (30 μg), quinupristin (15 μg), and linezolid (30 µg) (Mast Group, Merseyside, UK) was assessed by the Kirby-Bauer disk diffusion method on Mueller-Hinton agar (Merck, Germany) based on Clinical Laboratory Standards Institute (CLSI) criteria (8). Enterococcusfaecalis ATCC 29212 was used as the control strain (8).
According to CLSI criteria, vancomycin-sensitive Enterococcus (VSE) is defined as inhibition zone equal or greater than 17 mm. In addition, vancomycin-intermediate Enterococcus (VIE) and VRE are defined as the inhibition zone of 15 to 16 mm and equal or less than 14 mm, respectively.
3.3. DNA Extraction
Total DNA of the different bacterial isolates was extracted by the DNA extraction kit (Cat. No. K-3032-2, Bioneer Company, Korea). The process was performed as follows:
Two hundreds of phosphate buffer was added to the microtubes and colonies of bacteria were solved in microtubes; proteinase K (20 mL) and binding buffer (200 mL) were added to the microtubes. Then microtubes were placed in a water bath for ten minutes and elution buffer was placed in water bath at the same time. Thereafter, isopropanol (100 ml) was added to the microtubes. Microtubes content were transferred to the filtered microtubes and they were centrifuged. Microtubes content were emptied and washing buffer 1 (500 mL) was added and they were centrifuged. Microtubes content were emptied again and washing buffer 2 (500 mL) was added and they were centrifuged. Microtubes content were empty once more and they were centrifuged. Then elution buffer (100 mL) was added to the microtubes and they were centrifuged. Extracted DNA samples were transferred to new microtubes.
3.4. Detection of Virulence Genes by Polymerase Chain Reaction
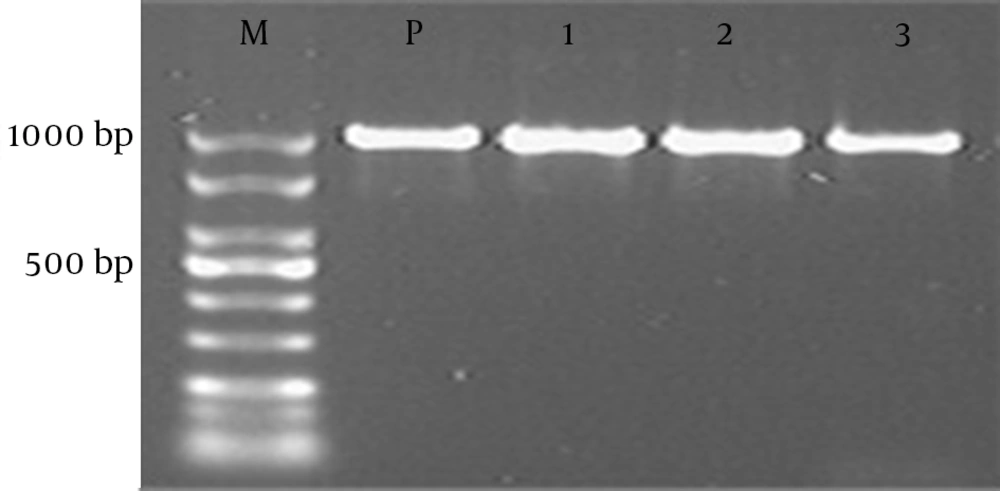
Polymerase chain reaction (PCR) method was performed on enterococcal isolates for detection of vanA and vanB genes. Used primers are presented in Table 1. Briefly, the 25 µL of PCR mixture contained 2.5 µL of bacterial DNA, 10 pM of each primers, 1.5 mM of MgCl2, 250 µM of each dNTP, 10 mM of Tris-HCL (pH = 9.0), 30 mM of KCL, and 1 U of Taq DNA polymerase (Bioneer Company-Korea, Cat. No. K-2012). Reactions were performed on thermal cycler (Eppendorf, Master cycler gradient). Amplification for vanA was performed with the following thermal cycling conditions: ten minutes at 95°C and 30 cycles of amplification consisting of one minute at 94°C, one minute at 57°C, one minute at 72°C, and ten minutes at 72°C for the final extension. Amplification for vanB was performed with the following thermal cycling conditions: 10 minutes at 95°C and 30 cycles of amplification consisting of one minute at 94°C, one minute at 60°C, one minute at 72°C, and ten minutes at 72°C for the final extension. PCR product bands were analyzed after electrophoresis on a 1% agarose gel at 100 V for 60 minutes in 1X TBE containing ethidium bromide and the result was checked under ultraviolet irradiation.
| Target | Product Length, bp | Company |
|---|---|---|
| vanA | 1030 | |
| vanA-F: 5ʹCATGAATAGAATAAAAGTTGCAATA3ʹ | Bioneer | |
| vanA-R: 5ʹCCCCTTTAACGCTAATACGATCAA3ʹ | Bioneer | |
| vanB | 433 | |
| vanB-F: 5ʹGTGACAAACCGGAGGCGAGGA3ʹ | Bioneer | |
| vanB-R: 5ʹCCGCCATCCTCCTGCAAAAAA3ʹ | Bioneer |
a Abbreviations: F, forward; R, reverse.
3.5. Sequencing Method
The PCR purification kit (Bioneer Co., Korea) was used to purify PCR products and sequencing was performed by the (Bioneer Company, Korea). The nucleotide sequences were analyzed with the Chromas 1.45 software and BLAST in NCBI.
4. Results
Seventy-one patients who met the inclusion criteria over a period of 18 months were enrolled in this study. Amongst them, 38 (53.5%) were males and 33 (46.5%) were females; mean age of patients was 29.1 ± 38.5 months (ranging from two days to 147.5 months). Sixty-four (90.1%) patients were colonized with enterococci. Of 64 strains, 47 (73.4%) were VRE. The remaining isolates were either VSE (11 strains, 17.2%) or VIE (six strains, 9.4%). Table 2 shows other demographic and clinical characteristics of the patients. The resistance rate of enterococci isolates to tested antibiotics were 85.11% to ampicillin, 80.85% to ciprofloxacin, 82.98% to penicillin, 87.23% to rifampin, 78.72% to teicoplanin, 46.81% to chloramphenicol, 23.40% to quinupristin and 2.13% to linezolid. PCR analysis of VRE samples showed that 42 (89.3%) samples had vanA gene but vanB gene was not detected in remaining samples. In VIE group, vanA gene was also detected in 4 (66.7%) cases. Again, we did not detect vanB gene in remaining samples (Figure 1).
| Characteristic | Positive for Growth of Enterococcus | VIE (n = 6) | VSE (n = 11) | VRE (n = 47) |
|---|---|---|---|---|
| Solid tumor | 3 (4.2) | - | - | 3 (4.2) |
| Diabetes mellitus | 2 (2.8) | - | 1 (1.4) | 1 (1.4) |
| Chronic renal disease | 4 (5.6) | 1 (1.4) | 2 (2.8) | 1 (1.4) |
| Treatment with chemotherapeutic agents | 8 (11.3) | - | - | 8 (11.3) |
| Blood dyscrasia | 8 (11.3) | - | - | 8 (11.3) |
| Chronic lung disease | 3 (4.2) | - | 1 (1.4) | 2 (2.8) |
| Immunodeficiency | 3 (4.2) | - | - | 3 (4.2) |
| Treatment with corticosteroids | 2 (2.8) | - | - | 2 (2.8) |
| Presence of invasive device | 44 (62) | 5 (7) | 8 (11.3) | 31 (43.7) |
| Previous ICU admission in the past 3 months | 11 (15.5) | - | 2 (2.8) | 9 (12.7) |
| previous treatment with antibiotics | 12 (16.9) | 2 (2.8) | 1 (1.4) | 9 (12.7) |
| ICU admission over 7 days | 47 (66.2) | 4 (5.6) | 7 (9.9) | 36 (50.7) |
a Abbreviations: VIE, vancomycin-intermediate Enterococcus; VRE, vancomycin-resistant Enterococcus; VSE, vancomycin-sensitive Enterococcus.
b Data are presented as No. (%).
5. Discussion
This study showed that vanA gene was the dominant resistance mechanism in isolated enterococcal colonization from patients admitted to ICU of this pediatric tertiary educational center in Tehran, Iran, during 2012-2013. Although many risk factors have been proposed to affect VRE colonization, assessment of these risk factors was not our primary goal in this study due to our small sample size. We used previous proposed risk factors to collect as much as possible VRE strain to look for resistance genes. However, it should be noted the among some of inclusion criteria, ICU admission over seven days, previous ICU admission in the past three months, presence of invasive devices, treatment with previous antibiotic, treatment with chemotherapeutic agents and having underlying hematologic problems seems to put patients more at risk of colonization (5, 9).
VRE colonization rates were investigated in various settings with inconsistent results; in a study in a tertiary care center in Australia, univariate analysis showed that the use of any antibiotic including meropenem as well as ciprofloxacin, diarrhea, and longer length of hospital stay were associated with increased risk of VRE colonization. The predominant VRE genotype circulating in Australia is E. faeciumvanB. In contrast, the vanA gene was predominant one in our study; however, our result was more compatible with VRE genotype status in United States and Europe (9). In another study in Italia, it was shown that 20 out of 26 VRE isolates from patients admitted to an ICU had vanA gene (10) while remainder showed vanB gene; a finding that was different from our results. Although remainder of our non-vanA VRE isolates might have less resistant genotypes, such a hypothesis must be investigated in future studies. Increasing enterococci with vanA gene rate in a hospital is a concerning issue for whole country. A study in Germany investigated the samples of different origin in various hospitals over the country between 2004-2006 and analyzed them by multilocus sequence typing (MLST), SmaI macrorestriction analysis in pulsed-field gel electrophoresis (PFGE), and multiple-locus variable-number tandem repeat analysis (MLVA) (11). A dissemination of related vancomycin-resistant E. faecium among various hospitals and Federal States was proved by spreading an identical vanA gene clusters among clonally different strain types. Hence, adherence to infection control measures especially in ICU settings is very important (11).
Our previous study in 2008 in Ali-Asghar Children Hospital, which was focused on VRE rectal colonization rate, detected VRE in stools from 33 (25%) of 130 children with acute lymphoblastic leukemia (ALL). No clear risk factors were identified for VRE colonization in that study, but there was a trend towards an increased prevalence in children admitted to the ICU since their ALL diagnosis (P = 0.07). The vanA gene was found in 28 (85%) of the 33 stools, with all other enterococci being vanB (12).
Although we assessed the rectal colonization in selected high-risk patients, the observed high rate in our study is concerning. Most studies in developed countries showed lower rates of colonization. In a study in Australia in a general hospital, VRE was detected from patients in each ward with the prevalence ranging from 3% to 29% and concluded that exposure to some antibiotics, especially meropenem, might explain the increasing rate of colonization. Therefore, the antimicrobial prescription in our setting should be closely monitored to prove their role in this high and increasing rate of colonization (9). Present study confirmed that ICU admission in our setting might be considered as a risk factor for VRE colonization and the fact that rate of colonization in selected cases was much higher in comparison with previous study. Moreover, the genotypes could be changed over time, as we did not detect any vanB gene in present study even in patients with malignancies. This finding necessitates further molecular studies to clarify the exact nature of resistant genes in these isolates.
Our study has some limitations; the sample size for most potentially risk factors was too low to do statistical analysis; hence, a multicentric national study is suggested to investigate the trend and exact risk factors in each hospital setting that would lead to design scientific preventive measures.