1. Background
Staphylococcus epidermidis is one of the main members of coagulase-negative staphylococci (CoNS), and may cause severe infections, such as bloodstream, medical implant devices, peripheral or central intravenous catheters (CVCs), Eye keratitis, and nosocomial infections (1, 2). Bloodstream infection is one of the most noticeable and common infections in many wards of health care units, especially in the intensive care unit (ICU) and neonatal intensive care unit (NICU) (3). The existence of virulence factors (biofilm formation, polysaccharide intercellular adhesion, phenol-soluble modulin and antibiotic resistance genes) and moreover the prevalence of resistant strains to many antimicrobial agents such as beta-lactams are important reasons of speared of infections in coagulase-negative related diseases. In the two past decades the incidence of Methicillin-resistant Staphylococcus epidermidis (MRSE) strains is becoming a global concern in ICU and other wards of hospitals (4, 5). The presence of the mecA gene is the main cause of MRSE emergence, and is related to a penicillin binding protein (PBP2a). The mecA, mecI and mecR1 are important elements of the mec operon, which are located on Staphylococcal Cassette Chromosome mec (SCCmec) (6). Up to now different types of SCCmec elements have been identified, however type I to V are most commonly found in Staphylococcus epidermidis strains (7). Since the SCCmec elements are classified on the basis of their various structural attributes, this kind of typing is considered a powerful method for MRSE epidemiological studies (8). On the other hand, Pulse Field Gel Electrophoresis (PFGE) is regarded as one of the gold standard methods for S. epidermidis epidemiological studies and is also useful for tracking specific outbreaks (9). It should be noted that the Multi-Locus Sequence Typing (MLST) method is another prominent typing technique for investigating many bacterial pathogens, which lead to nosocomial infections, including S. epidermidis (10).
2. Objectives
The aim of this study was to evaluate the molecular characterization of MRSE strains isolated from hospitalized patients in an ICU ward with SCCmec, PFGE and MLST typing methods.
3. Methods
3.1. Bacterial Isolates and Identification
In the six-month period of the study, 121 isolates were recovered from bloodstream infections of ICU hospitalized patients in Al-Zahra hospital (Isfahan, Iran). Among the collected isolates, 53 isolates were identified as Staphylococcus epidermidis and other isolates were Escherichia coli and Pseudomonas aeruginosa, after identification with primary microbiological tests (2). Staphylococcus epidermidis isolates were transferred to the microbiology laboratory and confirmed with microbiological standard tests such as Gram staining, catalase, coagulase activity testing with rabbit plasma, DNase, mannitol salt agar, and thereafter stored at -20°C for molecular supplementary tests.
3.2. Antimicrobial Resistance Profiles
Antibiogram was verified by disc diffusion method on Mueller-Hinton agar, according to Clinical and Laboratory Standards Institute (CLSI) guidelines (11). The antimicrobial agents included cefoxitin (30 µg), penicillin (10 IU), cefazolin (30 µg), vancomycin (30 µg), erythromycin (15 µg), gentamicin (10 µg), sulfamethoxazole (23.75 µg) + trimethoprim (1.25 µg), rifampicin (5 µg), levofloxacin (5 µg), clindamycin (2 µg) and teicoplanin (30 µg).
3.3. DNA Extraction
DNA extraction was performed using a DNA extraction kit (Qiagene, United States), according to the manufacturer's protocol. It should be mentioned that 1.5 μL of lysostaphin was added to the final bacterial suspension (12).
3.4. Determination of mecA Gene and SCCmec Types
Detection of mecA gene and various SCCmec types (I-V) was carried out by the multiplex polymerase chain reaction (PCR) approach with subsequent visualization of the amplified DNA fragment patterns by 1% agarose gel electrophoresis and SYBR® Safe DNA gel stain (Invitrogen, USA). Specific primers and PCR conditions were completed as described previously (12).
3.5. Pulsed Field Gel Electrophoresis
Methicillin-resistant Staphylococcus epidermidis strains were genotyped by Pulsed field gel electrophoresis (PFGE) using the restriction enzyme SmaI (Takara, Japan) and conducted according to methods described previously (13). Macrorestriction profiles were analyzed by GelCompar II version 4 (Applied Maths, Sint-Martens-Latem, Bel-gium) with 90% similarity cutoff point and clustered by unweighted pair group method with arithmetic mean (UPGMA).
3.6. Multi-Locus Sequence Typing
Polymerase chain reaction of the seven housekeeping genes encoding carbamate kinase (arcC), shikimate dehydrogenase (aroE), ABC transporter (gtr), DNA mismatch repair protein (mutS), pyrimidine operon regulatory protein (pyirR), triosephosphate isomerase (tpiA) and acetyl-CoA acetyltransferase (yqiL) was performed as explained previously (14). The allelic profiles and also sequence types were analysis with (http://www.mlst.net).
4. Results
During six months of the study, 121 bloodstream infections were collected from the ICU ward of Al-Zahra hospital. Fifty-three of all isolates belonged to S. epidermidis. The other covered isolates were associated with E. coli and P. aeruginosa. Since many factors were regarded for the determination of the clinical significance of coagulase negative staphylococcus diseases, in which the most prominent was center for disease control (CDC) standards, therefore all of the S. epidermidis isolates were gathered according to the mentioned guidelines (15). The antibiogram showed the following results: cefoxitin (43.4%), penicillin (100%), cefazolin (94.3%), vancomycin (0%), erythromycin (77.3%), gentamicin (73.6%), sulfamethoxazole/trimethoprim (56.5%), rifampicin (39.6%), levofloxacin (52.8%), clindamycin (43.4%) and teicoplanin (0%).
In the present study, 25 (47.2%) of S. epidermidis strains had the mecA gene. In addition, by multiplex PCR, SCCmec type II was detected in one (4%), SCCmec type III in eight (32%) and SCCmec type IV in thirteen (52%) of isolates. It is noteworthy to mention that three (12%) of the isolates were distinguished as non-typable and also SCCmec types I and V were not detected.
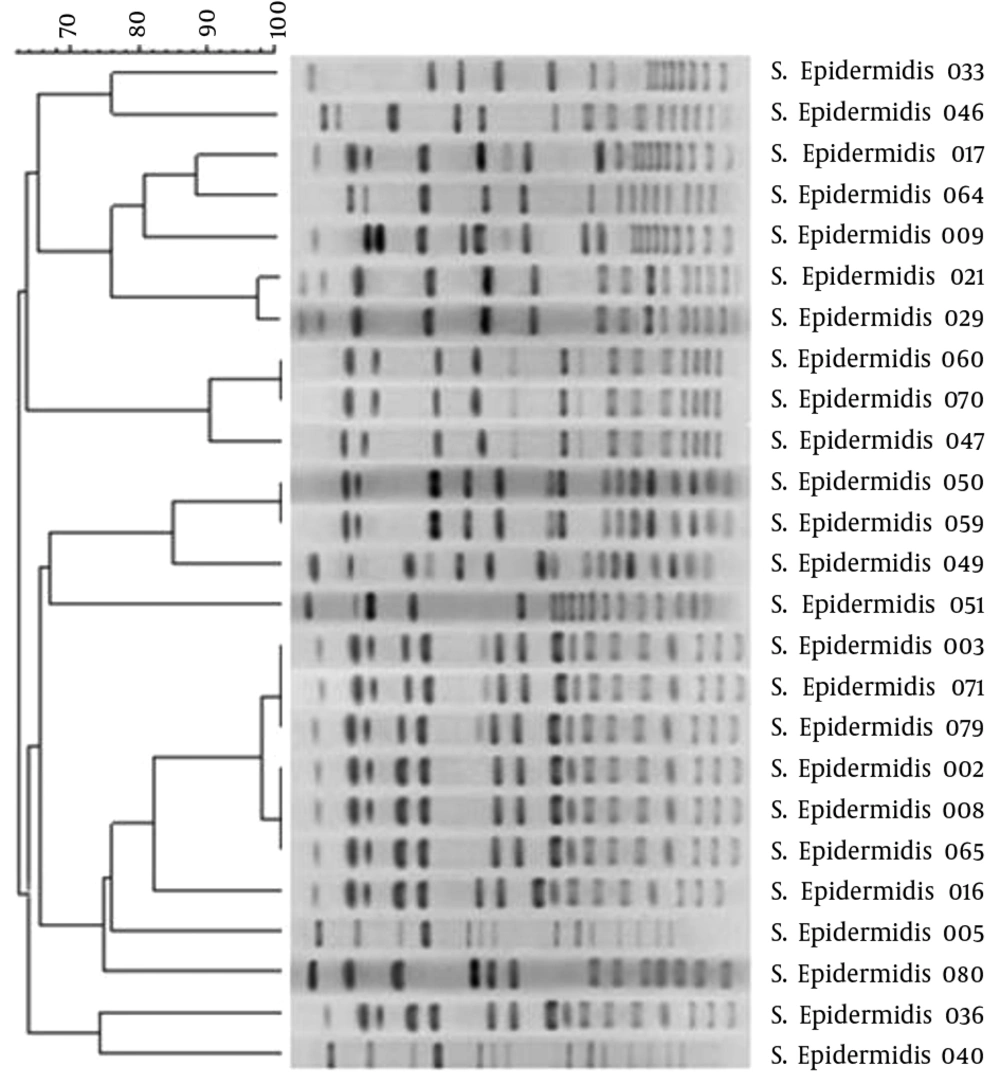
Using PFGE typing by GelCompar II analysis, a total of 16 different pulsotypes were identified, including four main clones (I-IV). In PFGE dendrogram clone I with two, clone II with three, clone III with two and clone IV with six strains had the most frequency (Figure 1).
Finally the MLST was carried out for I-IV detected clones by PFGE profiles. From clone I (strains 21 and 29), clone II (strains 47 and 60), clone III (strain 50 and 59) and clone IV (strains 3 and 8) were selected for MLST test, in accordance to SCCmec types. The MLST results are shown in Table 1.
| Sequence Type | No. (%) |
|---|---|
| ST2 | 5 (62.5) |
| ST5 | 2 (25) |
| ST27 | 1 (12.5) |
5. Discussion
According to many studies, nosocomial dissemination of S. epidermidis strains and the genetic diversity due to horizontal gene transferring and mobile genetic elements and exchanging may lead to different infections in various wards of health care units such as ICU (16). Currently researchers are aware that S. epidermidis and other (CoNS) are a reservoir of SCCmec elements and antimicrobial resistance genes.
The purpose of this study was to determine the SCCmec elements, PFGE pattern and the MLST of MRSE strains isolated from ICU wards. Antibiotic susceptibility testing results showed multidrug resistant isolates.
In the present study, the high antimicrobial resistance rates were evaluated in MRSE strains and 75.5% of the isolates were resistant to more than four antibiotics. In PCR molecular test, 47.2% of strains were distinguished as MRSE. It is important to note that no vancomycin and teicoplanin resistance were observed in our studied strains. Rifampicin with 39.6% resistance, had the lowest resistance among of antimicrobial tested agents.
In the study of Al Tayyar et al. 92 (41.2%) of the clinical specimens were isolated from the ICU ward. Of these, 69 (30.9%) of the specimens were associated with bloodstream infections. Furthermore, 54.7% of isolates belonged to S. epidermidis strains. In antimicrobial susceptibility test, vancomycin, linezolid, rifampin and nitrofurantoin showed high sensitivity while, the lowest sensitivity rate was distinguished in ampicillin, penicillin, ceftriaxone, cefazolin, amoxicillin-clavulanic acid and erythromycin (17).
In a similar study, which was done in Indonesia at the ICU ward, the main isolates were P. aeruginosa, K. pneumoniae and S. epidermidis. High rate of resistance to cephalexin and also levofloxacin was detected among S. epidermidis isolates (18).
In SCCmec typing, the prevalence of type IV of SCCmec was notable. These results were similar to Wisplinghoff et al. and other related studies (19). However in comparison to related studies about S. aureus isolates, the prevalence of SCCmec type III had the highest percentage (20, 21).
By the PFGE method, with 16 different pulsotypes, four main clones were obtained, amongst which, clone IV with six strains was the most common. This result showed the wide diversity of isolates by PFGE typing method. According to Table 2, most of the SCCmec results and PFGE patterns were coordinated with each others, for example, clone I or (F) had the same SCCmec type (IV) or clone IV or (K).
| Strains | Gender | Age | mecA gene | SCCmec Type | PFGE Profile | Allelic Profile | ST |
|---|---|---|---|---|---|---|---|
| 2 | M | 52 | Positive | Non-Typable | K2 | - | - |
| 3 | M | 63 | Positive | IV | K1 | 1-1-1-2-2-1-1 | ST5 |
| 5 | F | 45 | Positive | IV | M | - | - |
| 8 | M | 64 | Positive | IV | K2 | 1-2-2-2-2-3-1 | ST27 |
| 9 | M | 69 | Positive | III | E | - | - |
| 16 | M | 53 | Positive | IV | C | - | - |
| 17 | F | 50 | Positive | II | L | - | - |
| 21 | F | 49 | Positive | IV | F1 | 7-1-2-2-4-1-1 | ST2 |
| 29 | M | 18 | Positive | IV | F2 | 1-1-1-2-2-1-1 | ST5 |
| 33 | M | 68 | Positive | III | A | - | - |
| 36 | F | 49 | Positive | IV | O | - | - |
| 40 | M | 55 | Positive | III | P | - | - |
| 46 | F | 37 | Positive | IV | B | - | - |
| 47 | M | 77 | Positive | IV | G2 | 7-1-2-2-4-1-1 | ST2 |
| 49 | M | 49 | Positive | IV | I | - | - |
| 50 | M | 56 | Positive | III | H1 | 7-1-2-2-4-1-1 | ST2 |
| 51 | F | 68 | Positive | Non-Typable | J | - | - |
| 59 | M | 70 | Positive | IV | H1 | 7-1-2-2-4-1-1 | ST2 |
| 60 | M | 38 | Positive | III | G1 | 7-1-2-2-4-1-1 | ST2 |
| 64 | F | 67 | Positive | III | D | - | - |
| 65 | M | 53 | Positive | IV | K2 | - | - |
| 70 | M | 71 | Positive | III | G1 | - | - |
| 71 | F | 66 | Positive | IV | K1 | - | - |
| 79 | F | 76 | Positive | Non-Typable | K1 | - | - |
| 80 | M | 48 | Positive | III | N | - | - |
Multi-Locus Sequence Typing was performed on eight isolates, wherein three STs, including ST2, ST5 and ST27, were distinguished. ST2 showed the highest ratio in our studied isolates. From clone I (F1 and F2) each of the two strains were examined by MLST technique, while both strains had SCCmec type IV but the STs were different from each other (ST2 and ST5). From clone II (G1, G1 and G2) two strains (47 and 60) with the same STs were detected, while the SCCmec types were various. In clone III (H1 and H1) both strains (50 and 59) with different SCCmec types had the identical STs (ST2) and in clone IV with six strains, both strains number 3 (ST5) and 8 (ST27), were evaluated with MLST. In a related study, which was performed on blood cultures in Germany, 13 different STs were determined. The most common STs were allocated to sequence types ST2, ST5, ST10 and ST242. In our study, ST2 and ST5 had the highest prevalence sequence types, similar to the present study (22).
According to our research, the SCCmec typing, PFGE and MLST results had conformity with each other. To compare SCCmec and PFGE with established typing methods, we also analyzed eight of the S. epidermidis isolates by MLST. Taken alone within the studied S. epidermidis isolates, the PFGE data suggest a high degree of genotypic diversity; nevertheless these results have been verified by other investigations (23).
These great variations, which were observed using PFGE, suggest genetic diversity of various strains isolated from different patients, but in MLST a measure of genetic changes for housekeeping genes was involved in our targets so the rate of genetic diversity was happened slowly (24). This investigation indicated that from 53 S. epidermidis isolates, 47.2% were MRSE, which led to nosocomial infections in the ICU. On the other hand, the S. epidermidis with various SCCmec elements and resistance genes may lead to severe infections in health care units and accelerate nosocomial infections. The emergence of drug resistance amongst S. epidermidis strains is very critical point for nosocomial infections, especially for ICU and NICU patients, hence recognizing the source of infection is very essential to monitor the dissemination of infections, therefore this important issue is not addressed without various typing methods. According to antibiotic susceptibility test and molecular typing results, we suggest that it is better to evaluate important virulence factors of this microorganism because there may be a correlation between these results and virulent strains.