1. Background
Urinary tract infections (UTIs) are the most common human bacterial infections in both community and hospital settings for all ages (1, 2). It is estimated that every year about 150 million people are infected with UTIs at a cost of 6 billion US dollars worldwide (3). UTIs may involve the lower and sometimes both the upper and lower urinary tracts (4). In community-acquired UTIs (CA-UTIs), women are more likely to contract these infections in their life cycle than men (5, 6). The most common problem is recurrent infection which can affect women of all ages, especially pregnant and elderly women (7).
Both host factors and virulence factors released by the pathogens are involved in the pathogenesis of recurrent infections. Predisposing host factors to recurrent infections include genetic factors, aging, menopause, and urogenital dysfunction (7, 8).
Previous studies have shown that Escherichia coli are the most common organisms isolated from uncomplicated UTIs (4). However, enterococci, particularly Enterococcus faecalis, have been identified as second agents for CA-UTIs in some countries (9-12) and are reported to be responsible for 6 - 10% of CA-UTIs (13-16). This bacterium is a normal inhabitant of the gastrointestinal tract in animals and humans. However, it can also be an important pathogen that causes endocarditis, surgical wound infection, bacteraemia, and sepsis (17, 18).
The use of broad-spectrum antibiotics to treat UTIs can lead to an emergence of enterococci such as E. faecalis that are resistant to beta-lactam antibiotics, aminoglycosides, and glycopeptides, which is a major, global problem in the treatment of this illness (19-21).
The resistance patterns of E. faecalis in both urine and fecal specimens have not been studied extensively (22). In the past few years, the antibiotic resistance patterns of E. faecalis causing UTIs have changed in both the community and health care centers (23-25). This information is a reflection of changes over the years. A monitoring period appears to be necessary for the reduction of the number of UTIs (26-28). Tetracycline resistance in many commensal and pathogenic bacteria can be related to transferable genetic elements like plasmids (29). The vanA gene is one of the important causes of resistance to vancomycin and teicoplanin. The vanA gene can be located on a plasmid or transposon and can be spread among bacterial species rapidly (30).
Moreover, several studies have recorded that enterococcal infections are often caused by the patient’s own commensal flora (31). Determining the antibiotic resistance patterns of enterococci that have been isolated from both urine and feces and antibiotyping can be useful for discovering endogenous CA-UTIs. However, there is little data for antibiotic resistance patterns of E. faecalis in urine and fecal samples of outpatients with UTIs in Iran.
2. Objectives
The present study was designed and performed in Mofid hospital and Labbafinejad hospital to investigate the antibiotic resistance patterns among E. faecalis strains isolated from fecal and urine samples of patients with CA-UTIs using phenotypic and molecular methods to detect the vanA and tetM genes.
3. Materials and Methods
3.1. Isolate Collection and Identification
A descriptive study was conducted from August 2014 to March 2015 on outpatients attending Milad hospital and Labbafinejad hospital in Tehran, Iran. A total of 72 urine and fecal specimens were collected from consecutive outpatients with a UTI. An early morning midstream urine specimen was collected in a sterile container from these patients. Fresh fecal samples were also collected at the same time. Samples were transferred to the Pediatric Infections Research Center of Shahid Beheshti University of Medical Sciences at Mofid children’s hospital.
Urine specimens were inoculated on blood agar plates using calibrated loops and fecal specimens were inoculated on enterococcosel agar (BBL, USA) plates and incubated at 37°C for 24 hours. Colony forming units per milliliter numbering more than 105 was considered as bacteriuria. The colony morphology and culture characteristics were observed macroscopically. Standard biochemical procedures such as gram staining, the catalase test, the bile esculin hydrolysis test, growth in 6.5% sodium chloride, and the arabinose fermentation test were used for the isolation of E. faecalis strains (32, 33). All strains were stored at -70°C in trypticase soy broth with 20% glycerol. The PCR method was performed with primers specific for the E. faecalis species (see the Molecular examinations section).
3.2. Antibiotic Susceptibility Testing
The antibiotic susceptibility patterns of E. faecalis isolates in both samples were determined using the standard Kirby-Bauer disk diffusion method and the E. test according to the CLSI’s recommendations (CLSI 2014). After inoculation and preparing the disks, the Mueller-Hinton agar plates were incubated for 24 hours and the inhibition zones were measured with a metric ruler. A total of 12 antimicrobial agents were tested. These agents were penicillin G (10 units), ampicillin (10 µg), vancomycin (30 µg), tetracycline (30 µg), minocycline (30 µg), ciprofloxacin (5 µg), levofloxacin (5 µg), gatifloxacin (5 µg), nitrofurantoin (300 µg), gentamicin (120 µg) and linezolid (30 µg) (MAST GROUP Ltd., United Kingdom). MIC Test Strips (Liofilchem®, Italy) were used for detection of antimicrobial susceptibility to daptomycin. The E. faecalis strain ATCC 29212 and Staphylococcus aureus ATCC 25923 were used on each day of testing as quality controls. Multidrug resistance (MDR) was defined as resistance to three or more different classes of the antimicrobial agents tested.
3.3. Molecular Examinations
Extraction of the genomic DNA was performed using the High Pure PCR Template Preparation Kit (Roche, Germany), with some modifications. The bacterial pellet was mixed with 200µl PBS, digested in 5µl lysozyme, and incubated at 37°C for 15 minutes. The mixture was then lysed using a short incubation with a lysis buffer and proteinase k. The solution was then transferred to a spin column to remove any contaminating cellular components. Finally, the DNA was eluted using an elution buffer at 70°C.
PCR was carried out in a total volume of 25 μL Master mix 2x (Cinnagen, Iran) containing 50 pmol of primers, 100 ng of genomic DNA, 0.4 mmol L-1 of each of four dNTPs, 3 mmol L-1 MgCl2, and 0.08 U of Taq DNA polymerase. The primer sequences used were ddlE1 (ATCAAGTACAGTTAGTCTTTATTAG) and ddIE2 (ACGATTCAAAGCTAACTGAATCAGT) for E. faecalis isolates (32) with an amplicon size of 941 bp, vanA-F (5’-CATGAATAGAATAAAAGTTGCAATA-3’) and vanA-R (5’-CCCCTTTAACGCTAATACGATCAA-3’) with an amplicon size of 1030 bp (32), and tetM-F (5’-GGACAAAGGTACAACGAGGAC-3’) and tetM-R (5’- GGTCATCGTTTCCCTCTATTACC-3’) with an amplicon size of 446 bp (33). PCR was performed in a thermal cycler (Eppendorf, Germany) under the following conditions: initial denaturation step at 95°C for 5 minutes followed by 35 cycles consisting of denaturation at 95°C for 1 minute, annealing at 49°C (for ddlE), 57°C (for vanA), and 54°C (for tetM) for 1 minute, and 72°C for 1 minute followed by a final extension at 72°C for 10 minutes to ensure full extension of the PCR products.
The PCR amplification products were detected through electrophoresis in a 1% agarose gel followed by staining with red safe solution and a 100 bp DNA ladder (Fermentas, Germany). The results were visualized under a UV transilluminator. Internal positive controls have been used for the detection of tetM and vanA genes in this study. The direct sequencing of PCR amplified products was carried out using an ABI 3730X capillary sequencer (Genfanavaran, Macrogen, Seoul, Korea).
4. Results
4.1. Bacterial Isolation
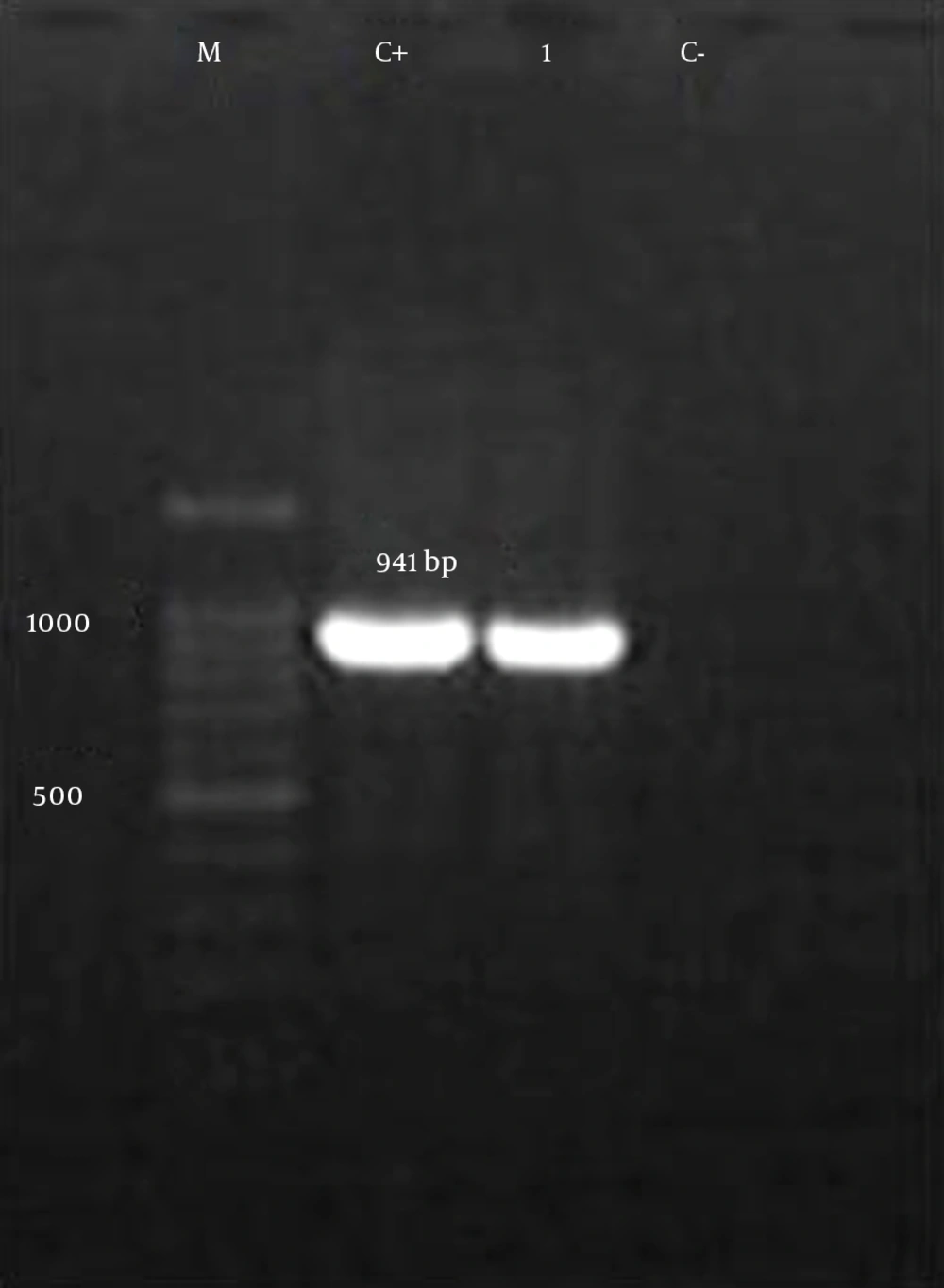
Of the 72 strains of E. faecalis isolated from the urine of patients with CA-UTI, 63 (87.5%) strains were also isolated from the feces of patients. Amplification of the E. faecalis-specific target produced a 941 bp band (Figure 1). The mean age for the studied group was 41.6 years, and the range was from 6 and 87 years. Thirty-seven isolates were collected from females (51.38%) and 35 from males (48.61%). In females, the majority of isolates came from patients who were 41 to 50 years old. In males, the majority of isolates came from patients who were 31 to 60 years old. The demographic characteristics of patients with CA-UTIs are shown in Table 1.
| Age Groups, y | Females, No. (%) | Males, No. (%) |
|---|---|---|
| 0 - 10 | 0 | 1 (2.85) |
| 11 - 20 | 4 (10.81) | 3 (8.57) |
| 21 - 30 | 5 (13.51) | 4 (11.42) |
| 31 - 40 | 8 (21.62) | 6 (17.14) |
| 41 - 50 | 11 (29.72) | 7 (20) |
| 51 - 60 | 7 (18.91) | 6 (17.14) |
| 61 - 70 | 2 (5.4) | 5 (14.28) |
| 71 - 80 | 0 | 1 (2.85) |
| 81 - 90 | 0 | 2 (5.71) |
| Total | 37 (51.38) | 35 (48.61) |
Patient Age and Sex Distribution
4.2. Antimicrobial Resistance
The susceptibility of E. faecalis isolates to various antibiotics in both urine and fecal samples is shown in Table 2. In general, the antibiotic resistance of strains isolated from urine was evaluated for tetracycline (65 strains [90.3%] resistant), minocycline (64 [88.9%]), gentamicin (120 µg) (21 [29.2%]), ciprofloxacin (17 [23.6%]), levofloxacin (12 [16.7%]), and gatifloxacin (11 [15.3%]). The same evaluation was conducted for the strains isolated from feces for tetracycline (48 [76.1%]), minocycline (45 [71.4%]), gentamicin (10 [15.8%]), ciprofloxacin (8 [12.6%]) and gatifloxacin (4 [6.3%]). In the present study we did not detect any resistance to vancomycin, ampicillin, penicillin, nitrofurantoin, or linezolid in isolates from urine or fecal specimens. All studied isolates in both samples were susceptible to daptomycin with a minimum inhibitory concentration (MIC) value of ≤ 4µg/mL. The number of strains with the MDR phenotype isolated from the 72 urine specimens was 9 (12.5%) and for the 63 fecal specimens was 7 (11.1%), as shown in Table 3.
| Antibiotic Disks | Urine Sample, N = 72 | Fecal Sample, N = 63 | ||||
|---|---|---|---|---|---|---|
| R, % | I, % | S, % | R, % | I, % | S, % | |
| Ampicillin | 0 | - | 100 | 0 | - | 100 |
| Penicillin G | 0 | - | 100 | 0 | - | 100 |
| Vancomycin | 0 | - | 100 | 0 | - | 100 |
| Linezolid | 0 | - | 100 | 0 | - | 100 |
| Nitrofurantoin | 0 | - | 100 | 0 | - | 100 |
| Gatifloxacin | 15.3 | - | 84.7 | 6.3 | - | 93.6 |
| Levofloxacin | 16.7 | - | 83.3 | 6.3 | - | 93.6 |
| Ciprofloxacin | 23.6 | - | 76.4 | 12.6 | - | 87.3 |
| Gentamycin (120 µg) | 29.2 | - | 70.8 | 15.8 | - | 84.1 |
| Minocycline | 88.9 | - | 11.1 | 71.4 | - | 28.5 |
| Tetracycline | 90.3 | - | 9.7 | 76.1 | - | 23.8 |
| Daptomycin | 0 | - | 100 | 0 | - | 100 |
Rate of Resistance and Susceptibility to Antimicrobial Resistance in E. faecalis Isolates from Urine and Fecal Specimens in CA-UTIs
| Antibiotic Resistance Patterns | Urine Sample, N = 72 | Fecal Sample, N = 63 |
|---|---|---|
| TET, MIN, GM120, CP, LEV, GAT | 7 (9.7) | 5 (7.9) |
| TET, MN, GM120, CP | 2 (2.7) | 2 (3.1) |
| TET, MN, GM120 | 11 (15.2) | 10 (15.8) |
| TET, MN, CP, LEV, GAT | 4 (5.5) | 3 (4.7) |
| TET, MN, CP | 3 (4.1) | 2 (3.1) |
| TET, MN | 37 (51.3) | 33 (52.3) |
| CP, LEV | 1 (1.3) | 1 (1.5) |
| TET | 1 (1.3) | 2 (3.1) |
| GM120 | 1 (1.3) | 0 |
Antibiotic Resistance Patterns in E. faecalis Isolates from Urine and Fecal Specimens in CA-UTIa
The most common MDR phenotype in strains isolated from both urine and fecal specimens was tetracycline/minocycline/gentamicin (120 µg)/ciprofloxacin/levofloxacin/gatifloxacin and for fecal specimens (Table 3).
In this study, 40 (63.4%) of the isolates from urine and fecal specimens shared similar antibiotic sensitivity and resistance patterns. 17 (26.9%) of the isolates in both samples were different in one class of antimicrobial agent that considered as related and 8 (12.6%) of the isolates in more than one or two classes of antimicrobial agents as difference. A similar correlation of antibiotic resistance patterns in these isolates is presented in Table 4.
| Antibiotic Resistance Patterns | Urine /Fecal Samples N = 63 |
|---|---|
| TET, MN, GM120, CP, GAT, LEV | 2 (3.7) |
| TET, MN, CP, GAT, LEV | 1 (1.5) |
| TET, MN, GM120 | 6 (9.5) |
| TET, MN | 25 (39.6) |
| TET, MN, CP | 1 (1.5) |
| LEV, CP | 1 (1.5) |
| TET | 1 (1.5) |
| Total | 37 |
Similar Correlation of Antibiotic Resistance Patterns in E. faecalis Isolates Between Urine and Fecal Samples from CA-UTI Patientsa
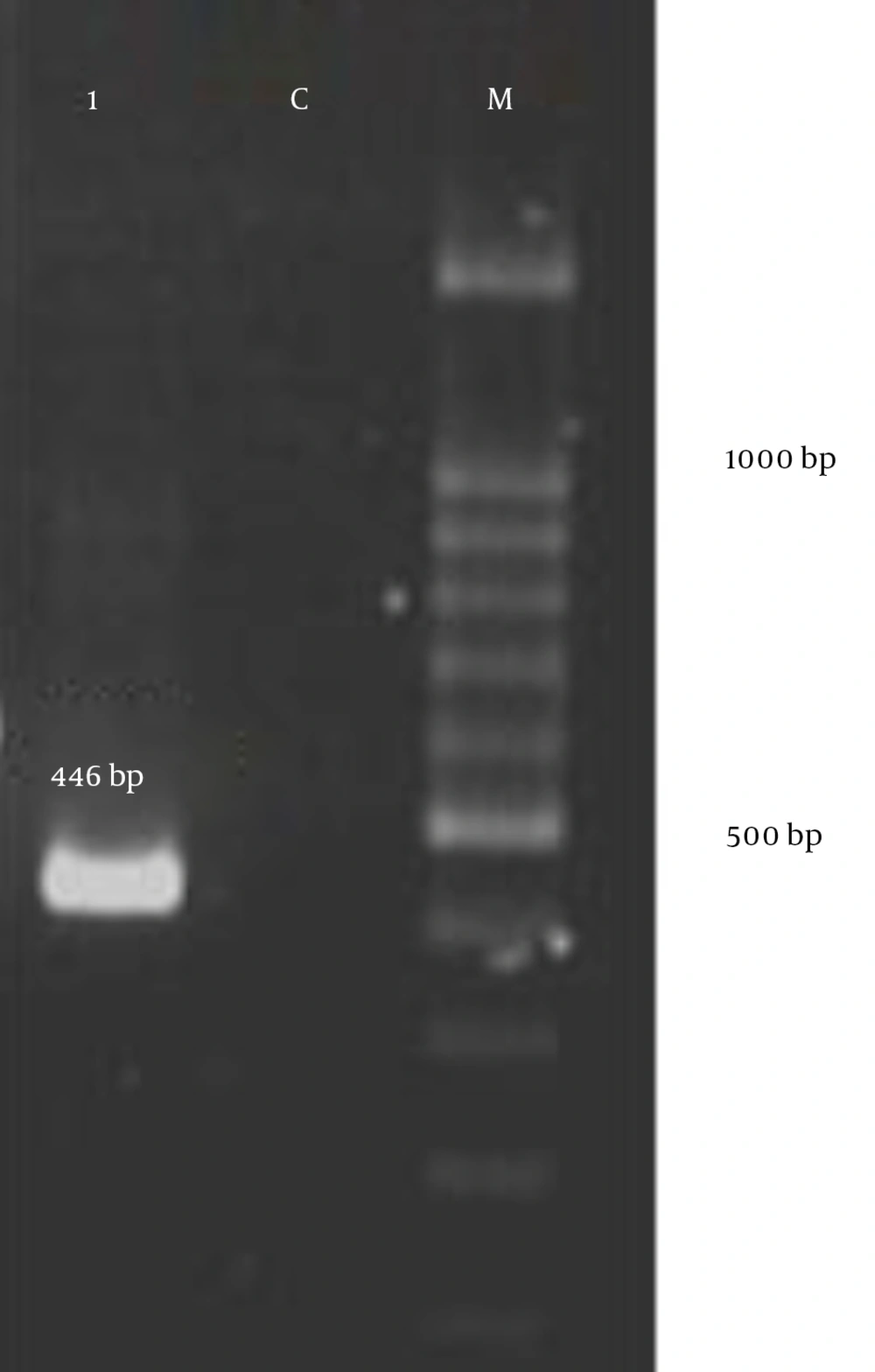
PCR results showed that the vanA gene was not found in any strain and 58 (92%) of the isolates from urine and 52 (82.5%) of the isolates from feces were positive for tetM genes. The amplification of tetM genes produced a 446 bp band (Figure 2). The sequencing pattern of the tetM gene confirmed the PCR results.
5. Discussion
UTIs represent one of the most common infectious diseases both in the community and hospital settings, and influence all age groups including men, women, and children around the world (34, 35). Enterococci, especially E. faecalis, have been considered as second agents for CA-UTIs in some countries (9, 10, 12). Our study showed a higher prevalence of CA-UTIs in females (51.38%) than in males (48.61%), which is similar to other findings (36-38).This high prevalence in women can be due to sexual intercourse, incontinence, and poor toilet hygiene (39-41).
In our study, all E. faecalis strains isolated from urine specimens were susceptible to penicillin G and ampicillin, which is lower than the 100% and 97.8% resistance rates reported in India (16), 57.1% in Iraq (15), and 59.8% in Portugal (42). Our results were comparable to the 0.02% and 0.04% resistance rates reported in Taiwan (43) and 0.3% in Brazil from urine specimens of patients with CA-UTIs (44). In this study, all isolates showed no resistance to linezolid and daptomycin, which is comparable to the 100% sensitivity rate from urine specimens reported in Taiwan (43). All of the strains were susceptible to nitrofurantoin, which is comparable to the 0.8% resistance rate from urine specimens reported in Brazil (44) and 100% sensitivity rate in India (16). In this study, vancomycin-resistant E. faecalis were not found in these samples which is comparable to the 100% sensitivity rate reported from urine specimens in Saudi Arabia (9) and 0.08% in Taiwan (43) and is higher than the 25% resistance rate reported from India (16). The reason for the absence of resistance to these antibiotics in this study and the low rates of resistance in other countries (43, 44) can be related to the fact that these antibiotics are not used as therapeutic antimicrobial agents in infections other than UTIs in Iran. Therefore, these antibiotics could be a good choice of antibiotic therapy for enterococcal CA-UTIs in Iran. On the other hand, high antibiotic resistance rates in other countries can be associated with the indiscriminative use of antibiotics. In this study, the vanA gene was not found in any strain. There are few studies about the prevalence of this gene in CA-UTIs around the world. A study in America and Canada was performed on inpatients and outpatients with UTIs, and 56.8% of the E. faecalis isolates displayed the vanA phenotype (45). The high prevalence of this gene could be due to the excessive use of vancomycin in these countries.
A low percentage of quinolone resistance was found in the strains studied here. The results from this study revealed that 23.6% of the E. faecalis strains from urine specimens were resistant to ciprofloxacin, which is comparable to the 25% resistance rate reported in India (16), lower than the 42.9% resistance rate reported in Iraq (15) and 38.1% in Portugal (42), and higher than the 9.7% resistance rate reported in Taiwan in CA-UTIs (46). The 16.7% resistance rate to levofloxacin in these isolates was higher than the 9.8% resistance rate reported in Taiwan (43).
Enterococci, including E. faecalis, have an intrinsic low-level resistance to aminoglycosides (46). In our study, the 29.9% resistance rate to gentamicin (120 µg) in these isolates from urine specimens was lower than the 50% and 42.9% resistance rates reported in Iraq and Taiwan, respectively (15, 43).
In our study, the majority of E. faecalis isolates from urine specimens were resistant to tetracycline (90.3%) and minocycline (88.9%), which is comparable to the 91.8% resistance rate reported in Taiwan (44) and lower than the 50% and 59.2% resistance rates reported in India and Brazil, respectively (16, 43). The resistance gene tetM that mediates resistance through ribosomal protection was detected in 92% of the urine specimens in the current study. The prevalence of this gene in the present study is significantly higher compared to other studies. In a study conducted in China by Jia et al., the prevalence of the tetM gene was reported in 31.6% of the E. faecalis strains (47). No information has been reported about the frequency of this gene in CA-UTIs in Iran. The high rate of tetracycline resistance in E. faecalis isolates may be related to the indiscriminate use of antibiotics in these patients and animal agriculture in Iran (48). Therefore, surveillance of the use of antibiotics in the community and surveys of animal reservoirs of tetracycline-resistant E. faecalis are essential (49).
There is little information about the MDR phenotype of E. faecalis isolates in CA-UTIs around the world. In this study, the percentage of the MDR phenotype was found to be 12.5% in E. faecalis isolated from urine specimens, which is lower than the 30% resistance rate from outpatients reported in Taiwan (43). No information has been reported about the frequency of the MDR phenotype in these isolates in Iran. The low prevalence of multiple antibiotic-resistant strains may be due to the large population of bacterial isolates which have not been exposed to several antibiotics.
Information is scarce about the antibiotic resistance of E. faecalis isolated from fecal specimens of patients with CA-UTIs. In a study conducted in Ethiopia on the antibiotic resistance of Enterococcus species isolated from the intestinal tracts of hospitalized patients, it was revealed that a high rate of fecal colonization by vancomycin-resistant enterococci was due to the use of vancomycin in hospitalized patients (48).
In this study, 63.4% of the isolates from urine and fecal specimens have similar antibiotic sensitivity and resistance patterns. This suggests the involvement of uropathogenic E. faecalis in the infection of these patients. The colonization of the gastrointestinal tract with different strains of E. faecalis or contamination was possibly responsible for the UTI in these patients. Further studies are essential to identify virulence factors involved in the colonization of these isolates and to determine the clonal relatedness of these strains using molecular fingerprinting methods in the urinary tract.