1. Background
Enteropathogenic Escherichia coli (EPEC) is a major cause of diarrhea among infants in developing countries (1, 2). The EPEC pathogenesis is based on an intimate adherence of bacteria to the intestinal epithelium cells, leading to the development of lesions called "attaching and effacing" (A/E) lesions (2, 3). The locus of enterocyte effacement (LEE) pathogenicity island harbors virulence factor genes, responsible for A/E lesions (2, 4). The LEE also encodes the type III secretion system (T3SS) that injects virulence factors to epithelium cells and changes the cytoscleton actin polymerization (5). The eaeA gene is located in the LEE and mediates intimate adherence of EPEC to the translocated intimin receptor (Tir) (6).
EPEC is divided into two groups, typical EPEC (tEPEC) and atypical EPEC (aEPEC). The adherence factor plasmid (pEAF) is present in tEPEC strains while being absent in aEPEC. The pEAF encodes the bundle forming pilus (BFP), a type IV pilus which contributes primary adherence of bacteria to epithelium cells (7, 8). In tEPEC, the BFP by localized adherence (LA) pattern attach to HeLa and HEp-2 cell surfaces. In contrast, the aEPEC strains form a loose attachment that is called localized like adherence (LA like) (9).
Traditionally, EPEC strains have been identified as E. coli serotypes that are epidemiologically related to children’s diarrhea. These strains are usually not identified in routine microbiology laboratories. According to many false results and low specifity of serotyping, many of the nonclassical EPEC serogroups were ignored (10, 11). Additionally, cell culture is an expensive and time-consuming method for distinguishing tEPEC from aEPEC strains.
Novel methods for detection and differentiation of E. coli pathotypes, especially EPEC strains, are based on polymerase chain reaction (PCR) (12). The EPEC strains have been identified by the presence of the eae gene and absence of the stx gene, which distinguish them from Entroheamorrhagic E. coli (EHEC). The tEPEC strains are differentiated from aEPEC by the presence of a bfpA gene (13, 14).
Random Amplified Polymorphic DNA is the PCR-based typing method which is mainly used to differentiate between different strains within the same species. In this method, one or more short primers were used to bind to various sites on the template DNA. The PCR yields a series of products with various sizes. The band pattern represents a genetic fingerprint characterizing a particular bacterial strain (15).
2. Objectives
In this study, we aimed to detect EPEC in E. coli isolates from children with and without diarrhea who were submitted to selected hospitals in Tehran from 2014 to 2015.
3. Methods
3.1. Sampling and Culture
In this descriptive study, from June 2014 to January 2015, total 140 and 110 E. coli isolates from children under five years old with and without diarrhea, respectively, were evaluated for the presence of EPEC. Samples were collected from Emam Hossein, Taleghani, and Loghman hospitals. In this study, diarrhea is defined as excretion of watery, loose, or mucuidal stool three or more times in one day. The samples were cultured on Macconkey and EMB agar (Merck, Germany) and were incubated at a temperature of 37°C overnight. Lactose positive colonies were examined with IMViC to identify E. coli isolates. Diarrhea samples were checked for WBC, RBC, and microscopic situation; corresponding patients were evaluated for abdominal pain, fever, and vomiting (15).
3.2. PCR Assay
The E. coli isolates were extracted for DNA with the boiling method (15). The isolates were confirmed as E. coli by positive PCR results of uidA (housekeeping beta D-glucuronidase gene). The E. coli K12 was used as a positive control. The confirmed isolates were examined for the presence of eae and stx genes, and the eae positive isolates were checked for presence of the bfpA gene. PCR amplification was performed in a 25 µL reaction mixture containing 2 μL DNA template, 12 μL ready to use Mastermix (2X) (Fermentase, Germany), 9 μL of distilled water, and 1 μL of 20 pmols forward and reverse primers. Positive control for eae and bfpA was EPEC E2348/69 and EHEC O157H7 used as positive control for the stx gene. The PCR conditions are mentioned in Table 1.
| Gene | Sequence (5' - 3') | Product Size, bp | PCR Conditions | Ref |
|---|---|---|---|---|
| uidA | 5-GCGTCTGTTGACTGGCAGGTGGTGG-3; 5- GTTGCCCGCTTCGAAACCAATGCCT-3 | 510 | {95°(5 minutes), 95°(30 seconds), 67°(30 seconds), 72°(30 seconds), 72°(5 minutes)} × 30 | (16) |
| eae | 5-CTGAACGGCGATTACGCGAA-3; 5-CGAGACGATACGATCCAG-3 | 918 | {95°(5 minutes), 95°(30 seconds), 58°(30 seconds), 72°(30 seconds), 72°(5 minutes)} × 30 | (17) |
| stx | 5-GAGCGAAATAATTTATATGTG-3; 5-TGATGATGGCAATTCAGTAT-3 | 518 | {95°(5 minutes), 95°(30 seconds), 52°(30 seconds), 72°(30 seconds), 72°(5 minutes)} × 30 | (17) |
| bfpA | 5-AATGGTGCTTGCGCTTGCTGC-3; 5-GCCGCTTTATCCAACCTGGTA-3 | 324 | {95°(5 minutes), 95°(30 seconds), 62°(30 seconds), 72°(30 seconds), 72°(5 minutes)} × 30 | (17) |
| RAPD | 5-CCGCAGCCAA-3 | - | {95°(5 minutes), 95°(30 seconds), 36°(90 seconds), 72°(60 seconds), 72°(5 minutes)} × 35 | (18) |
Primer Sequences, Sizes of Product Fragments, and PCR Conditions
3.3. RAPD-PCR
The genetic diversity of EPEC isolates was evaluated by RAPD-PCR. This assay was performed on all EPEC isolates by a decameric primer under conditions that are mentioned in Table 1. The different RAPD pattern is equivalent to genetically different isolates (18).
Statistical analysis: Chi square and Fisher’s exact tests were used for analysis of categorical data (sex, age, WBC, RBC, abbominal pain, and fever). Analysis was performed using Sigma Stat for Windows version 16 (SPSS, Chicago, IL, USA). A P value less than 0.05 was statistically significant.
4. Results
In this study, the mean age of the patients was 2.5 years. In patients with diarrhea, 31 (22.1%) patients had abdominal pain, 13 (12.1%) had fever, and 4 (2.8%) had vomiting; however, in patients without diarrhea just 2 (1.8%) had abdominal pain and 3 (2.7%) had fever. Also in diarrheal samples, PMNs and RBCs were seen in 38 (27.1%) and 2 (1.4%) samples, respectively, while in non-diarrheal samples were not seen with WBC and RBC. Five patients that were colonized with EPEC had WBC in stool and experienced abdominal pain and fever, and one patient had vomiting as well.
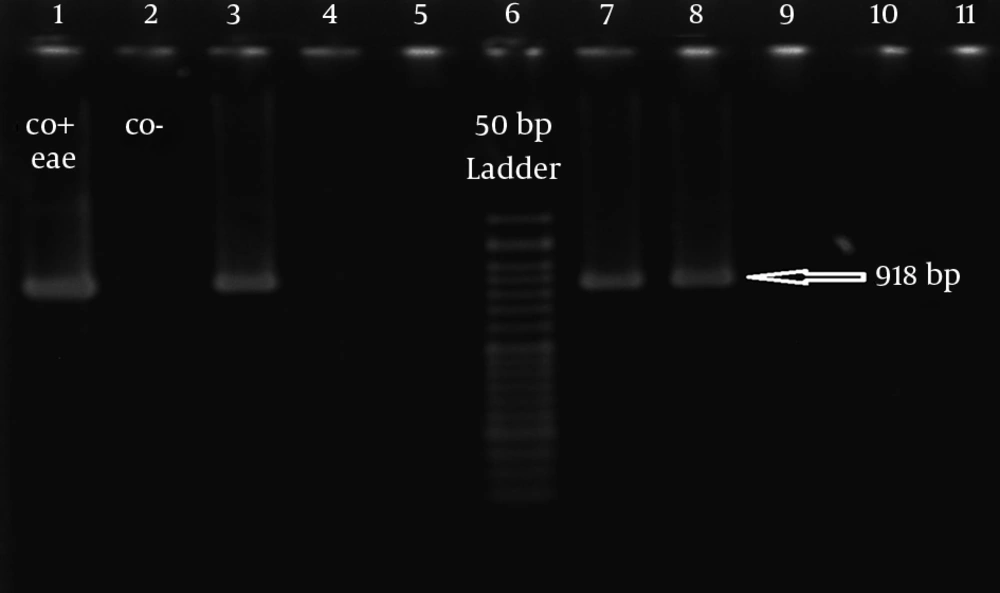
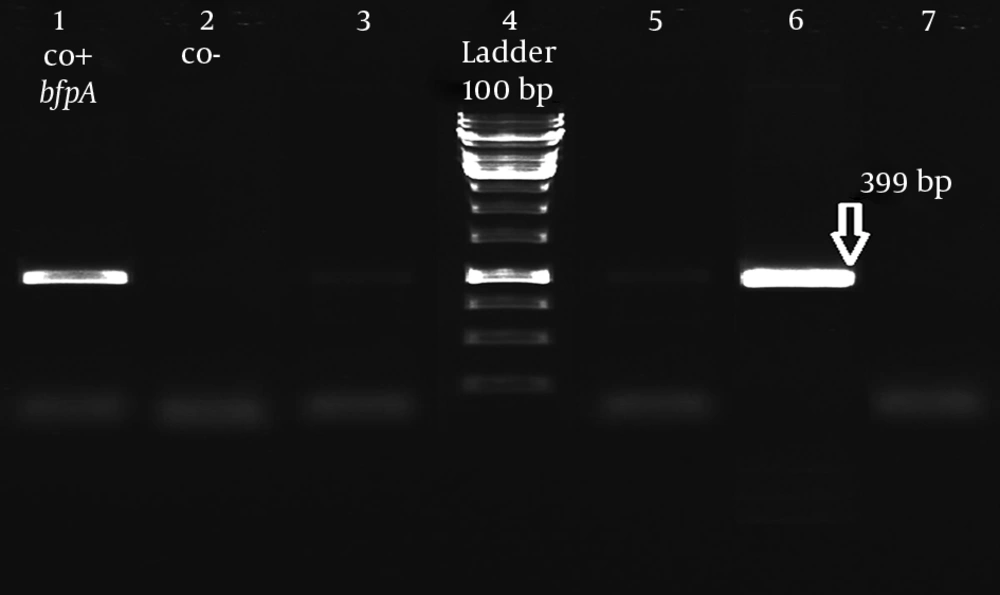
In isolates, all 140 diarrheal and 110 non-diarrheal E. coli isolates were confirmed by IMViC biochemical tests and positive PCR results of the uidA gene. Among 140 diarrheal isolates, 6 (4.28%) were positive for eae while all non-diarrheal isolates were negative. All 6 eae positive isolates did not harbor stx gene and identified as EPEC (Figure 1). According to bfpA amplification, 5 (83.3%) and 1 (16.7%) isolates identified as tEPEC and aEPEC, respectively (Figure 2). These results showed significant correlation of EPEC with diarrhea (P < 0.05). Significant correlation was also observed between EPEC isolation and the presence of WBC in stool, abdominal pain, and fever (P < 0.05).
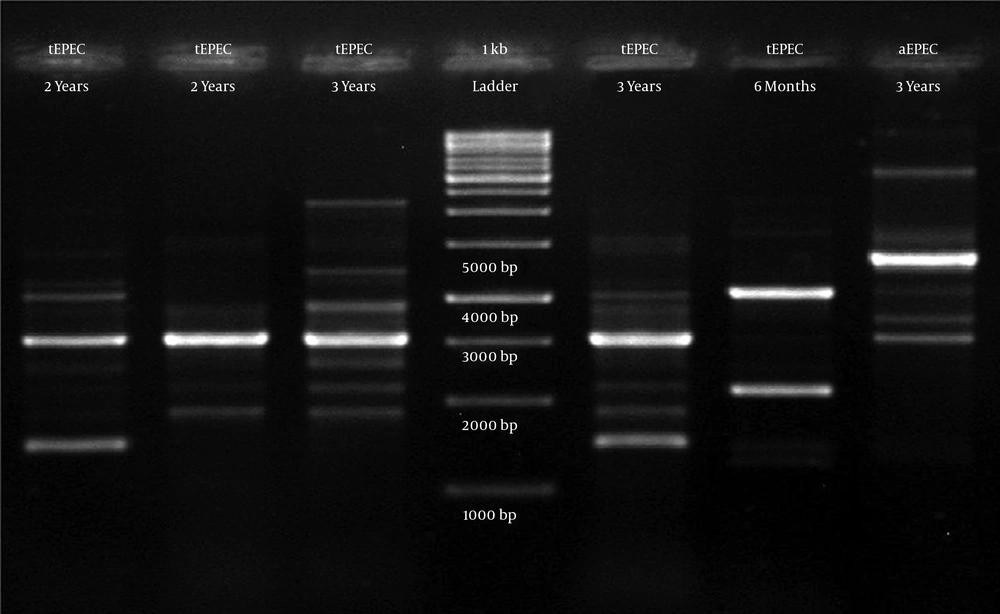
In RAPD-PCR, among 6 EPEC isolates, the 3 kb band was present in 5 isolates. Other bands of all 6 EPEC isolates were different. The range of band sizes was 1 to 10 kb. All 6 EPEC isolates showed a totally different pattern, and no similar types were seen (Figure 3).
5. Discussion
EPEC is a major cause of diarrhea among infants in developing countries. Traditional methods for distinguishing EPEC strains are expensive and time consuming, and in most cases they are incapable of differing between pathogen E. coli and normal flora. For this reason, there is no adequate information about the presence of these pathotypes as a cause of infant diarrhea.
Estrada-Garcia et al. (19) demonstrated a significant association of EPEC with acute diarrhea during 7 to 12 days among Mexican children, suggesting that EPEC is more associated with protracted diarrhea than the other diarrheagenic E. coli pathotypes. In our study, a total of 140 E. coli isolates were analyzed, of which 4.28% isolates were EPEC. The frequency of EPEC in other studies in Turkey and Iran were lower than our findings (10.3% and 38.4%, respectively) (14, 20). In 2009, Usein et al. detected EPEC in 36% of Romanian infants under five years old. All isolates in this study were categorized as aEPEC strains, while in recent study tEPEC strains were dominant (21).
In our study, the highest rate of EPEC was found in children between two and three years. Our statistical analysis suggests that EPEC was significantly associated with diarrhea in children in these two age groups (P < 0.05). This trend agrees with several studies, which have shown that the peak of enteritis was always in the few months after the beginning of the weaning period and that most EPEC infections occur in the first three years of life (22). However, other studies demonstrated that the incidence of community-acquired EPEC infection is highest in the six-month period following childbirth, and the infection is more severe in younger children (23, 24).
The majority of patients with diarrhea, which was infected with EPEC, were referred to hospitals during the summer. This is comparable to other studies (22, 25), which reported that EPEC infections are associated with the warm season (25); the peak rates of EPEC infection occurred mainly in the dry summer months. However, the differences were not found to be statistically significant.
In 6 patients with acute diarrhea, EPEC was isolated as the only pathogen, which demonstrates the importance of EPEC as the leading cause of infectious diarrhea.
All isolates in our study showed a diverse genetic pattern by RAPD analysis, which explain that these isolates were derived from various contamination sources.
Bando et al. used RAPD analysis for evaluating 73 strains of diarrheagenic E. coli isolates, and their results showed two divergent groups: one is genetically homogeneous EPEC whereas the other contains EPEC and non-EPEC isolates (26).
The RAPD analysis was also performed by George et al. for determination of genetic diversity of 352 E. coli isolates from urinary tract infection (UTI) in India. Their phylogenetic analysis showed five different subtypes (27).
In another study, Dulguer et al., 78 typical and atypical strains of EPEC and non-EPEC isolated from children with and without diarrhea were typed by RAPD-PCR. They demonstrated both typical and atypical strains are genetically distinct. They also found distinct types among isolates, which were collected from different regions (28).
The RAPD was used for differentiation of E. coli isolates from different sources in the prior reviewed studies. All of them showed E. coli isolates were totally genetically diverse. Those studies in aim, region, source of isolates, and RAPD primers, were different. The current survey was more comparable to research of Dulguer et al. in Brazil (28). Because of the low rate of EPEC isolation in this study, we could not analyze them by a dendrogram.
This study showed the significance and association of the strains of E. coli as a predominant isolates in diarrhea in children younger than five years in Tehran, Iran. These data suggested EPEC strains from different contamination sources are an important cause of diarrhea in Iranian children.