1. Background
There are increasing data regarding the roles of the intestinal microbiota in the pathogenesis of several human diseases (1). Prolonged interaction of these bacteria with the intestine or their overgrowth seems to be responsible for chronic diseases in this organ (2). Even in low numbers, pathogenic bacteria which are not common members of the gut microbiota can cause different human illnesses under some conditions (3, 4). Invasion of these pathogens into the intestinal barrier layer or the production of metabolites that dysregulate the normal signaling pathways of the intestinal cells is involved in the disease progression in infected patients. More than 200 transmitted microbial agents from foodstuffs are associated with gastroenteritis in the human population (5). Among these agents, the most important enteric bacterial pathogens that are responsible for gastrointestinal diseases are Escherichia coli, Campylobacter jejuni, Shigella spp., Salmonella spp., Vibrio cholera, and Yersinia spp. (6). Clostridium difficile is also considered the most frequently identified enteric pathogen in hospitalized patients with a recent history of medication (7). The infectious dose of these pathogens varies depending on their virulence potency and level of resistance to the harsh conditions of the gut environment (8). In some cases, gastrointestinal infectious diseases are acquired by the consumption of contaminated foods or water that are infected with fewer than 10 microorganisms (9). Detection of responsible bacterial agents in environmental or fecal samples mainly depends on the validity of the laboratory tests used and the sampling procedure, including time of sampling and transport conditions. It is hard to resolve challenges that exist for the detection and enumeration of these bacteria, since the causative bacteria are often present in low numbers within these samples or their presence is influenced by food materials or high counts of indigenous bacteria (10). Therefore, a rapid yet sensitive and specific diagnostic assay would be advantageous to clinicians for the early recognition of disease and to infection control practitioners for the swift implementation of control measures (11).
Bacterial culture is considered the “gold standard” for identification of diarrheagenic bacteria from stool specimens. However, this method is time consuming and laborious, requiring prolonged incubation, selective enrichment, and reduction of the background flora, which should be followed by biochemical identification tests (12, 13). Most laboratories are unable to diagnose the anaerobic, microaerophilic, and fastidious bacteria responsible for human gastrointestinal disorders using conventional microbiological methods. Recently, more rapid DNA-based methods for direct identification and even subtyping of these pathogens in stool specimens have been developed (14-21). Polymerase chain reaction (PCR) is a powerful technique for the detection of the target DNA in various clinical specimens, including fecal samples. However, fecal specimens often contain substances that may interfere with the PCR assay, leading to false-positive or false-negative results (2, 4, 11, 13, 15, 22). Accordingly, its application in clinical laboratories needs validation. Comparison of results for conventional culture- and molecular-based methods, which are designed for each bacterial species, is essential.
2. Objectives
In this study, we aimed to compare the detection limit and performance of PCR and culture methods for diagnosis of Campylobacter spp., Yersinia spp., C. perfringens, and C. difficile. As enteric pathogens, these are among most sensitive to the ambient culture conditions commonly used for diagnosis in clinical laboratories.
3. Methods
3.1. Bacterial Strains and Growth Conditions
The strains used in this study were the reference strains of Campylobacter jejuni (ATCC strain 33560), C. difficile (research center of gastroenterology and liver disease [RIGLD]-141), Yersinia enterocolitica (ATCC strain 101776), and C. perfringens (; RIGLD-2). Selective culture media, including Brucella agar (Merck, Germany; supplemented with 5% sheep blood and Campylobacter selective supplement), Clostridium difficile agar (Mast, UK; supplemented with defibrinated horse blood and Clostridium difficile selectavial), Yersinia selective agar (Merck; supplemented with Yersinia selective supplement CIN), and Egg Yolk agar (Merck; supplemented with neomycin), were used for the subculture of these strains. In the cases of C. perfringens and C. difficile, the cultures were incubated under anaerobic conditions (Anoxomat, MART Microbiology B.V.; 0% O2, 10% H2, 10% CO2, and 80% N2) at 37°C for 48 hours. Campylobacter jejuni was grown under microaerobic conditions (6% O2, 6% CO2, 3% H2, and 85% N2) for 24 hours at 42°C; Y. enterocolitica was grown under ambient air conditions at 25°C.
3.2. Spiked Stool Experiments
Serial tenfold dilutions of each bacterial species were freshly prepared at defined concentration (101 - 108 CFU/mL) in a control stool specimen and phosphate-buffered saline (PBS). A total of 100 µL of each dilution was spread onto the selective media for enumeration of the colony counts. In the cases of C. difficile, the inoculated samples were initially treated with alcohol and yeast extract broth to remove the common intestinal microbial flora. For alcohol treatment, about 1 g of stool was mixed with an equal volume of 95% methanol and then slowly vortexed and held at room temperature for 2 minutes. The treated suspensions were cultured on the selective media supplemented with 5% horse blood (CDSA). For the enrichment of Clostridium, nearly 1 g of the stool samples were mixed with an equal volume of yeast extract broth (Yeast extract; Merck). The treated suspensions were then cultured on the selective medium. To detect C. perfringens spores, the methanol (95%) and heat-treated spiked samples (90°C, 20 minutes) were cultured on Neomycin Egg Yolk agar. Cold enrichment in PBS and direct culture of the inoculated stool samples on CIN (agar 25°C) and MacConkey agar (37°C) were used for isolation of Y. enterocolitica. Two replicas of the tests were performed for analysis of the variations in our results (23).
3.3. DNA Extraction and PCR
DNA was extracted from the prepared stools according to the manufacturer’s instructions using the DNA Stool Kit (Bioneer, South Korea). The concentration of DNA samples was 30 µg. All of the DNA extracts were stored at -20°C until use. Amplification of target genes for the detection of Campylobacter spp. (16S rRNA), Yersinia enterocolitica (ompF), C. difficile (Cdd-3), and C. perfringens (16S rRNA) was performed using the specific primers depicted in Table 1. All PCR amplifications were performed in 25-μL volumes containing 3 μL of DNA template, 0.5-mM concentrations of deoxynucleoside triphosphates, 2.5 μL of 10 × PCR buffer (gene fanavaran), 0.75 mM MgCl2, 0.3 μM concentrations of each forward and reverse primer, 0.2 U of Taq DNA polymerase (Gene Fanavaran, Iran) under the following conditions: initial denaturation step at 95°C for 5 minutes, followed by 35 cycles of denaturation at 94°C for 1 minute, annealing for 1 minute, and extension at 72°C for 1 minute. After the last cycle, the mixture was incubated at 72°C for 5 minutes. The amplification products were analyzed by electrophoresis on a 1.5% agarose gel. Images were obtained after staining of the gels with ethidium bromide and under an ultraviolet (UV) light imaging system.
| Bacterium | Target Gene | Primer | Sequence (5´ to 3´) | Amplicon Size (bp) | Tm°C | Reference |
|---|---|---|---|---|---|---|
| Campylobacter spp. | 16S rRNA | C412F; C1228R | F: GGATGACACTTTTCGGAGC | 816 | 48 | (24) |
| R: CATTGTAGCACGTGTGTC | ||||||
| Yersinia spp. | 16S rRNA | 227Fmod; 669R | F: GTCTGGGCTTTGCTGGTC | 428 - 465 | 43 | (25) |
| R: GCGTCGTATTTAGCACCAACG | ||||||
| C. difficile | Cdd-3 | Tim6; Struppi6 | F: TCC AATAATAAATTAGCATTCC | 622 | 54 | (26) |
| R: GGCTATACACGTAATCCAGATA | ||||||
| C. perfringens | 16S rRNA | 184-205; 441-462 | F: AAAGATGGCATCATCATTCAAC | 279 | 50 | (27) |
| R: TACCGTCATTATCTTCCCCAAA |
| Limit of Detection, CFU/Gram Stool | ||||
|---|---|---|---|---|
| C. jejuni | C. difficile | C. perfringens | Y. enterocolitica | |
| Culture | 104 | 10 | 2 × 104 | 10 |
| PCR | 102 | 102 | 2 × 104 | 104 |
4. Results
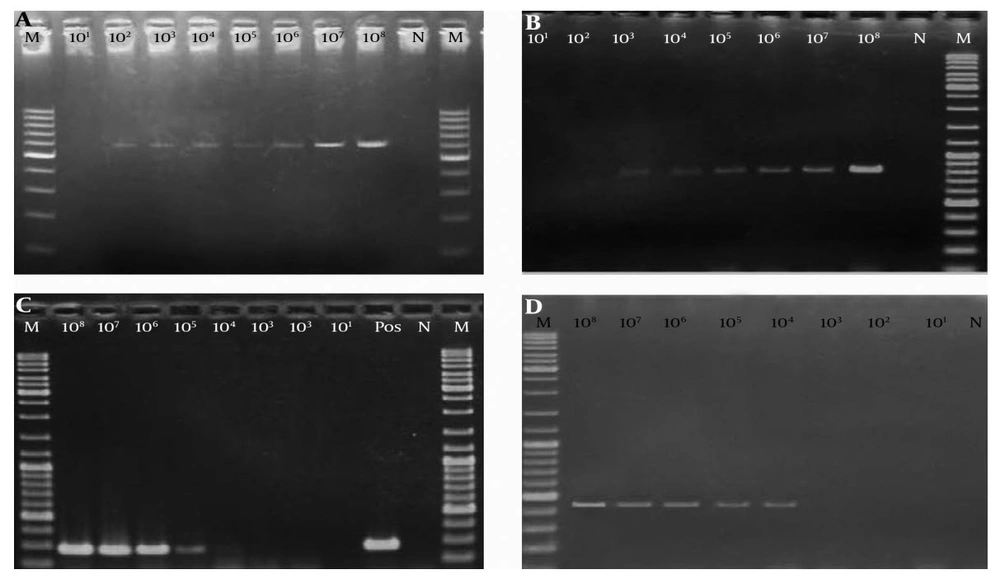
The results of the PCR assays showed specificity of primer pairs that were used for detection of the target bacteria. Accordingly, the species-specific primers provide a single PCR amplicon in the spiked stool samples (Figure 1). Analysis of culture method results for the detection of C. jejuni showed a sensitivity limit of 104 CFU/g for the spiked stool samples. This detection level was as low as 101 CFU when the bacterium was directly inoculated on the culture medium from the PBS suspensions. PCR results showed a lower detection limit for the spiked stool samples (102 CFU/g). The validity of these results was confirmed by obtaining the same results for both of the studied strains in two separate tests. Analysis of the culture results for detection of C. difficile on selective medium showed its sensitivity for the detection of 10 organisms in 1 gram of stool sample. The PCR detection limit for this bacterium was 100 CFU/g; however, an increased detection limit of up to 10 genome copies per PCR reaction was obtained on prepared dilutions of genomic DNA. In the case of Y. enterocolitica, there was also discordance between results obtained by culture and those using the PCR method. In the culture method, Y. enterocolitica was detected in all dilutions of bacteria in PBS. In contrast, a lower limit of detection was observed via PCR (104 CFU/g). The results of anaerobic culture for C. perfringens showed a detection limit of 2 × 104 CFU/g that was similar to those were obtained by PCR assay results.
5. Discussion
The prevalence levels of Campylobacter, Y. enterocolitica, and C. difficile are about 10.8%, 1.2%, and 21%, respectively, in stool samples of patients with gastroenteritis in developing countries (28-30). These rates are lower than those reported for developed countries (4, 31). Despite differences in geographic area and culture in these countries, inaccurate results could be obtained due to the need for individual equipment and defined culture media for the detection of these pathogens in patients’ stool samples. Because of their advantages, molecular tests are now widely used to detect these bacteria in clinical specimens. These tests are extremely useful diagnostic tools and are particularly valuable for the detection of infectious agents that are difficult to grow in conventional culture media. However, the presence of PCR inhibitors and variation of procedures that are used for sample preparation or extraction of nucleic acids affects their accuracy, precision, specificity, and sensitivity (23). The interpretation of results for these assays should be based on their limit of detection.
In this study, analysis of culture and PCR methods for the detection of C. jejuni showed a sensitivity limit of 104 and 102 CFU/g, respectively, for the spiked stool samples. In a study by Singh et al. (32), higher sensitivity of PCR compared with culture method was indicated in the spiked fecal samples (Table 3). Our results showed lower detection limits for both the culture and PCR assays compared with those reported by Persson et al. (33) (pure culture, 101-2 CFU; spiked stool, 105 CFU/g). The obtained detection level for conventional PCR in our study was similar to those reported for C. jejuni and C. coli using the real-time PCR method (2.5 × 102 CFU/g of feces) (34). The incongruent results could be explained as relating to differences in the primer sequences and DNA extraction methods used in this study. Recovery of C. jejuni from stool samples may be affected by the types of culture media used. Potturi-Venkata et al. (35) showed a higher isolation rate for modified charcoal cefoperazone deoxycholate (mCCDA) compared with Brucella agar–based media for isolation of Campylobacter spp. from fecal samples (Table 3). C. jejuni is a microaerophilic bacterium that requires specific incubation conditions to grow in synthetic culture media. Although usage of an effective culture medium will improve the isolation rate of the bacterium, the need to provide microaerophilic conditions for its growth and supplements to prevent the growth of fecal microbiota are considered the main limitations of this method.
| Bacterium | Method | Results, CFU g-1/DNA Copya | References |
|---|---|---|---|
| Campylobacter spp. | Culture and PCR | 104/102 | This study |
| Culture and PCR | 105/102 | (33) | |
| Real-time PCR | 2.5 × 102 | (36) | |
| qPCR | 102 | (37) | |
| Y. enterocolitica | Culture and PCR | 10/104 | This study |
| Multiplex PCR | 105 | (38) | |
| Real-time PCR | 102 | (39) | |
| Culture and PCR | 4 × 103/4 × 102 | (40) | |
| C. difficile | Culture and PCR | 10/102 | This study |
| Real-time PCR | 5 × 104 | (41) | |
| C. perfringens | Culture and PCR | 2 × 104/2 × 104 | This study |
| Multiplex PCR | 102-4 | (42) |
aCFU/g, colony forming unit/gram stool; DNA copy of target bacterium per gram stool was represented for all the molecular assays.
Alcohol pretreatment of stool specimens together with appropriate incubation time (up to 1 week) seems to be an effective method for the detection of C. difficile spores in the stool samples (7, 8); however, usage of other sensitive and rapid tests is preferable. In our experiment, the PCR detection limit for this bacterium was 100 CFU/g. It seems that anaerobic culture has a much lower detection limit (10 CFU/g) than PCR assay for the detection of C. difficile. Belanger et al. (41) used real-time PCR assay for detection of C. difficile, and their detection limit was estimated to be as high as 5 × 104 CFU per gram of feces. Using the prepared dilutions of genomic DNA, an increased detection limit of up to 10 genome copies per PCR reaction was obtained in our experiment. According to these results, the incongruence of the PCR results compared with the culture results could be explained by the low yield of DNA that was extracted from the C. difficile spores in the spiked stool samples or the existence of mutations in the cdd3 locus in the regions where our primers adhered. The existence of mutations was not supported by our data, since cdd3 was detected in the DNA extracts of diluted DNA samples at the lower concentration. In a study by Luna et al. (43), where tcdA and tcdB were targeted in the spiked stool samples by real-time PCR, the lower limit of detection of C. difficile was 250 CFU/mL for tcdA and 500 CFU/ml for tcdB. These researchers similarly concluded that the sensitivity of the tests can only be increased in the more concentrated samples.
Discordance between results of culture and PCR methods was also determined in the case of Y. enterocolitica. This finding was supported by Weimer et al. (38), who used multiplex real-time PCR for the simultaneous detection of Y. enterocolitica and other bacteria in stool samples; they reported a sensitivity limit of 105 CFU/g in their research. In another study, detection limits of 102 CFU/mL and 103 CFU/g for the pure culture and stool sample, respectively, were obtained using real-time PCR (39). However, Boyapalle et al. (40) reported a lower detection limit for PCR (4 × 102 CFU/g) compared with culture method (4 × 103 CFU/g). The lower sensitivity of the PCR method compared with the culture method was also confirmed by the results of our assay using DNA extracts of the provided dilutions of Y. enterocolitica in PBS (71.4%, 103 CFU/mL). In a study by Wannet et al. (44), those primers that targeted ail and 16s rRNA genes showed a sensitivity of 100% (one genomic copy) in pure culture of Y. enterocolitica. Differences in the primers that target the ompF gene for PCR and the type of DNA extraction kit used in our experiment may explain these contradictory results. Re-analysis of the tests using different primers and extraction kits will provide more accurate data about their limits of detections.
C. perfringens is not only a member of human microbiota in the gastrointestinal tract, but it is also considered as a common cause of food poisoning in foodborne outbreaks (45). In general, detection of > 106-8 CFU per gram of this bacterium in stool samples of patients with gastroenteritis is considered clinically important (42). Our results showed a similar detection limit (2 × 104 CFU/g) for both the culture and PCR methods. This amount was similar to those reported by Wise et al. (42) using a multiplex PCR assay. In their study, C. perfringens alpha and enterotoxin genes were targeted for detection of the bacterium in spiked fecal samples of domestic animals, and an average sensitivity of 102-4 CFU/g was reported. Our results illustrated a correlation between the PCR assay and traditional culture method for the detection of C. perfringens in fecal spiked samples. Since the studied samples were subjected to heat treatment for the elimination of non-spore-forming bacteria (which may affect the germinating cells of C. perfringens), it seems that the detection limits of these tests are lower than 103 CFU/g in human fecal samples.
The PCR results for all bacteria mentioned in this study were available on the same day as the assays were performed, whereas the culture results took 24 hours for Y. enterocolitica and 48 - 72 hours for C.jejuni, C. difficile, and C. perfringens. These results collectively showed that direct plating can be used successfully for the detection of anaerobic enteric bacterial pathogens (C. difficile and C. perfringens) and fastidious bacteria (Yersinia spp. and Campylobacter spp.) in human fecal samples when a bacterial load of greater than 104 CFU/gram is present. The specified PCR assays showed acceptable results with respect to detection limits, which makes these methods especially suitable for rapid diagnostics of slow-growing bacteria in the fecal samples of infected patients. Improvements in the DNA extraction method and target sequences of the primers are needed to achieve more accurate results.