1. Background
Streptococcus pneumoniae, an important Gram-positive pathogen, is responsible for infectious diseases such as meningitis, pneumonia, otitis media, and septicemia (1-3). These clinical forms are more common among young and elderly populations due to their lack of immune response to pneumococcal polysaccharide vaccines (4, 5). In such patients, the incidence of pneumococcal diseases caused by non-vaccine serotypes is increasing; a fact that highlights the importance of developing effective pneumococcal vaccines (6). Epidemiological studies also indicated that the incidence of invasive pneumococcal diseases is increasing, despite the availability of conjugate vaccines (7-10). In the United States, S. pneumoniae is responsible for 61% of meningitis cases, while in the developing countries, invasive pneumococcal diseases are estimated to cause 0.7 to 1.0 million deaths annually among young children (11). Therefore, rapid detection of organisms is important regarding the initiation of treatment. Conventional culture of respiratory secretions, including sputum, pleural, or bronchoalveolar lavage, is used to diagnose pneumococcal pneumonia and other lower respiratory tract infections; however, the yields are low and the method is time-consuming (12). In contrast with conventional techniques, molecular approaches to detect and differentiate microorganisms mostly depend on the amplification of DNA and are not affected by low microbial loads (13, 14). In addition, molecular procedures can be applied to organisms that are slowly growing in the culture (15). Advancement of molecular assays lead to the introduction of novel methods with enhanced sensitivity and specificity. A loop-mediated isothermal amplification (LAMP) assay is a diagnostic tool that amplifies DNA under isothermal conditions (3). In this assay, a set of 4 specific primers are annealed to 6 separate regions within the target sequence. The use of 4 primers (F3, B3, FIP, and BIP) results in increased efficiency and specificity (16). The loop primers increase the performance and rapidity of the LAMP method and can reduce the amplification time (17). Production of magnesium pyrophosphate results in white precipitate; therefore, the assay can be observed by the naked eye without any special processing or gel electrophoresis (18). Additionally, the LAMP assay is not affected by inhibitors remaining after the DNA extraction step. Therefore, relatively simple DNA extraction procedures can be used instead of commercial DNA extraction kits (19, 20). Pneumococci are genetically and metabolically similar to some mitis groups of streptococci, which reside in the human oral cavity and even can present commensal organisms (21). Based on the fact that differentiation of S. pneumoniae from mitis group streptococci is a key objective in clinical laboratories, development of a rapid and accurate assay is necessary to detect pneumococci.
2. Objectives
The differentiation of S. pneumoniae, as a pathogenic streptococcus, from mitis groups of streptococci is important. In the current study, a LAMP assay was used to target the choline-binding protein M (cbpM) as a conserved gene and its sensitivity was evaluated using 30 pneumococcal isolates.
3. Methods
3.1. Primer Design
The LAMP primers were designed using Primer explorer V4 (https://primerexplorer.jp/lamp4), deploying the software’s default settings to target cbpM gene. The LytA-specific primers were also designed using Primer3 software. The primer sequences are shown in Table 1.
| Name | Sequence (5’ → 3’) |
|---|---|
| SPF3 | GAGAGGGACGAGAGAACC |
| SPB3 | TTGAGGTAGTACCATGAACC |
| SPBIP | AGTTGCTCCAACAACAGGCTGGAAACAAGTACCCAACCTGTTGCCAT |
| SPFIP | CATCTAACCAAGGAACAGATGTAGCAGTAGAACAAAAGTTGTTGTTCCTGAGC |
| lytA (PCR) | F: ATGGAAATTAATGTGAGTAAATTAAGAA |
| R: TTTTACTGTAATCAAGCCATCTG |
3.2. Bacterial Culture and DNA Extraction
Streptococcus pneumoniae ATCC 49619, as well as 30 isolates of pneumococci (isolated from patients hospitalized at Tehran hospitals, Iran) and non-pneumococcal species were cultured on Trypticase soy agar (TSA) supplemented with 5% sheep blood, before incubation for 24 hours at 37°C in a 5% CO2 incubator. DNA extraction was performed using a DNA extraction kit (Promega Wizard Genomic DNA purification kit, Madison, WII, USA), based on the manufacturer’s instructions.
3.3. Optimization of LAMP Assay
The LAMP reaction mixture was prepared using a total volume of 25 µl containing 2.5 µL of 10X Bsm Buffer, 20 mM Tris-HCl (pH 8.8 at 25°C), 10 mM KCl, 10 mM (NH4)2SO4, 5 mM MgSO4, 0.1% Tween 20 (Thermo Scientific-Fermentas, Vilnius, Lithuania), different ratios of outer primers: inner primers, different concentrations of dNTPs (0.4, 0.6, 0.8, 1, and 1.2 mM), different concentrations of MgSO4 (2, 4, 6, 8, 10, and 12 mM), 8 U Bsm DNA polymerase large fragments (Thermo Scientific-Fermentas, Vilnius, Lithuania), and 1 μL of DNA template. The reaction mixture was incubated in a Thermoblock at various temperatures for several minutes and the products were finally electrophoresed on 3% agarose gel.
3.4. Specificity of LAMP and PCR Primers
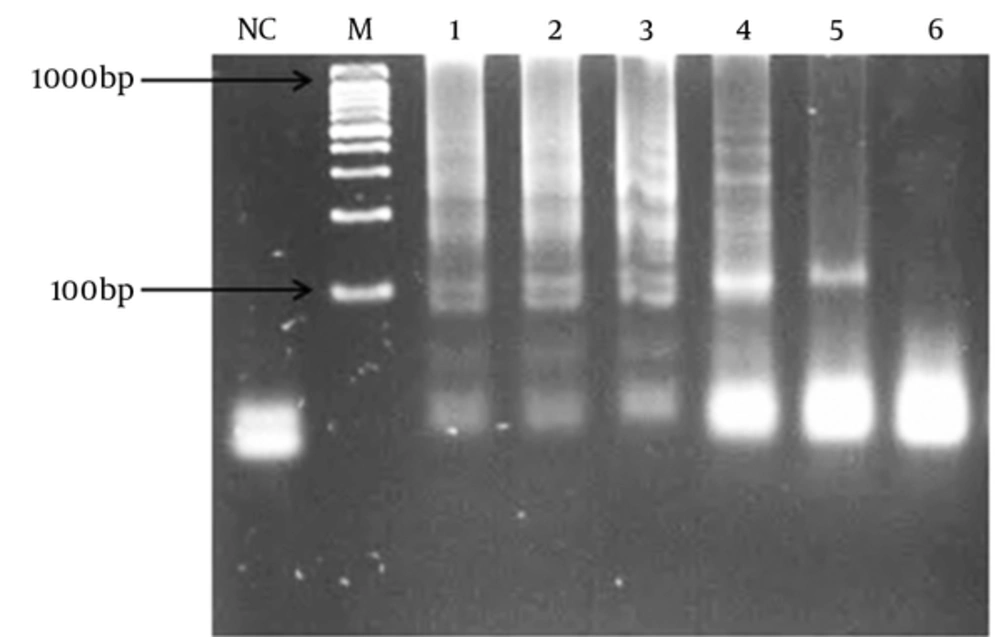
The specificity of the LAMP and PCR assays was evaluated using Streptococcus pyogenes ATCC 8668, S. mutans ATCC 35668, S. agalactiae ATCC 12386, S. sanguinis ATCC 10556, S. mitis ATCC 6249, S. dysgalactiae ATCC 35666, and Staphylococcus aureus. The LAMP and PCR products were analyzed by gel electrophoresis.
3.5. Sensitivity and Determination of Detection Limits of LAMP and lytA-PCR Assays
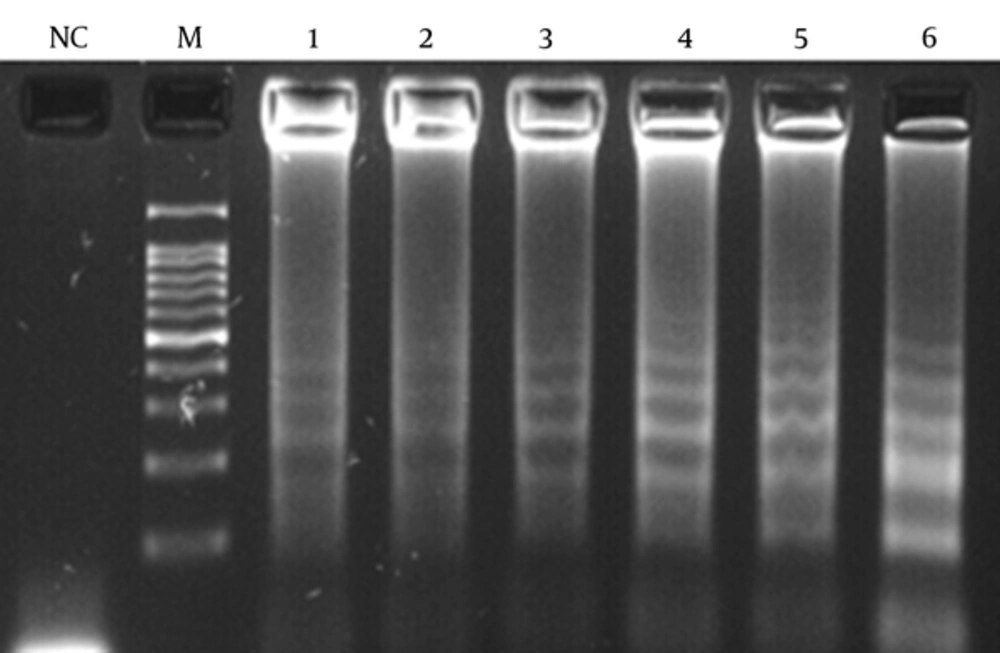
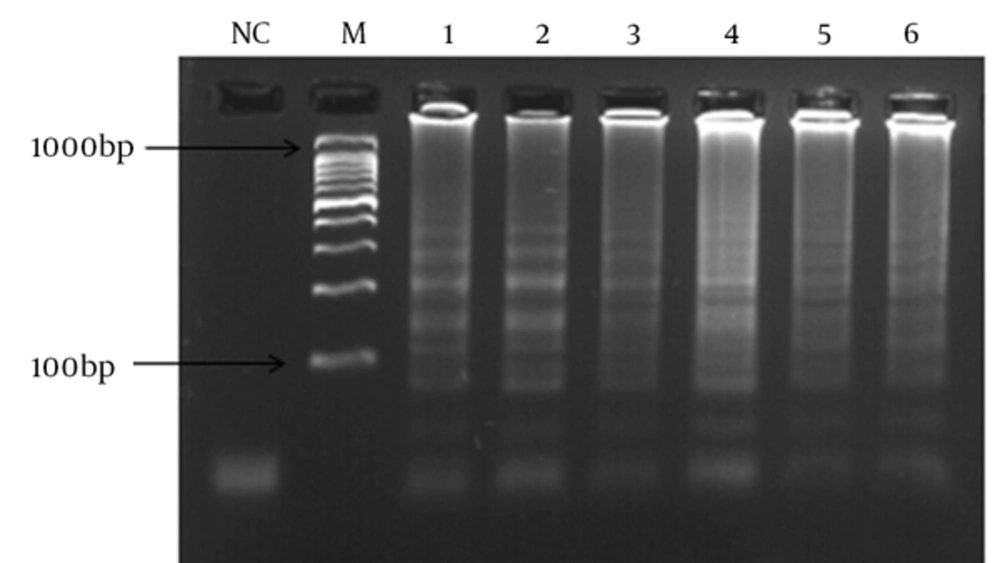
A 10-fold serial dilution of the DNA was prepared in sterile water, and the LAMP and PCR assays were carried out using different DNA concentrations. The sensitivity of the 2 assays was also evaluated by 30 pneumococci isolates.
4. Results
In all, 30 isolates of S. pneumoniae as well as 7 non-pneumococcal species (including Streptococcus pyogenes, S. mutans, S. agalactiae, S. sanguinis, S. mitis, S. dysgalactiae, and Staphylococcus aureus) were used in the current study. All species were cultured on TSA, and DNA extraction was conducted successfully.
4.1. Optimization of LAMP Assay
As described in the materials and methods section, the present LAMP assay was performed without loop primers. Pure DNA of S. pneumoniae ATCC 49619 was used as a template to determine the optimal conditions for the assay.
To optimize and find an optimal incubation temperature for LAMP assay, a standard reaction was carried out under a range of temperatures (60, 61, 62, 63, 64, and 65°C) for 60 minutes; maximum yield was achieved at 62°C. Therefore, the best temperature for the LAMP assay was established as 62°C used for all subsequent applications.
To denote the best period of time during which LAMP reaction amplification could yield sufficient observable products, 5 different times limits were applied for the said reaction; good results were obtained at 60 minutes.
Different MgSO4, dNTPs, and outer and inner primer concentrations were evaluated and the best results were obtained at 8, 0.4, and 1.6 mM, respectively.
4.2. Specificity of LAMP and PCR Primer
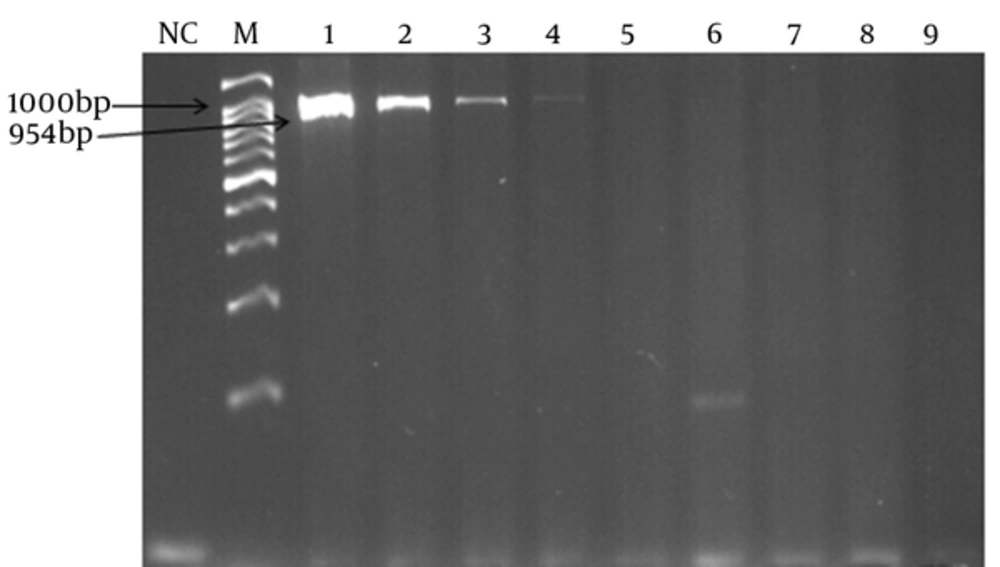
The specificity of the PCR and LAMP primers was tested using DNA samples from the 7 non-pneumococcal species. No amplifications were observed indicating specific primers.
4.3. Sensitivity and Detection Limit of Assays
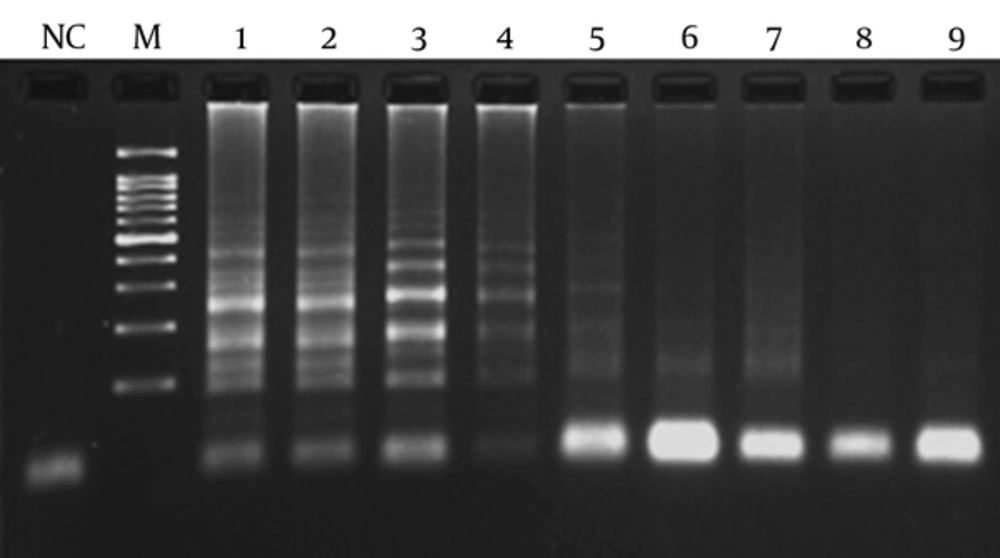
In order to determine the lowest detection limit of LAMP and PCR (using lytA primers), a 10-fold serial dilution (ranging from 10-1 to 10-9 of DNA) was provided. The results of amplification showed that the minimum copy number of DNA detectable by LAMP and PCR assays was 5 and 50, respectively. Both of the LAMP and PCR assays were positive when tested on the 30 pneumococcal isolates, thus the sensitivity of these assays was 100% using the 2x2 table of sensitivity.
5. Discussion
Streptococcus pneumoniae is a lethal human pathogen associated with high morbidity and mortality and leading to a global economic burden (22). As the spectrum of pneumococcal diseases is increasing annually, rapid detection of this organism is essential. Currently, there are multiple traditional assays to detect S. pneumoniae from various clinical specimens (23-25). However, the conventional procedures are time-consuming and not cost-effective. In the current study, a LAMP assay was developed to detect S. pneumoniae targeting cbpM. In several studies, autolysin genes (such as lytA) are used as the identification target (26-29). Although the lytA is conserved in all pneumococcal species (30), blasting the sequence shows similarity among the related sequences of mitis group streptococci. The current study applied the basic local alignment search tool (BLAST) to identify and select a specific conserved gene in all pneumococcal species, the results showing that cbpM is conserved in all S. pneumoniae species, but not in any of the mitis groups or other streptococci. The results of the LAMP and PCR assays also showed that all of the S. pneumoniae and non-pneumococcal isolates were positive and negative for the cbpM, respectively. Therefore, this small 387-bp gene can be useful in specific detection of S. pneumoniae. Moreover, lytA can also be an acceptable target to detect pneumococci, although the BLAST results showed its sequence similarity with some sequences of mitis group streptococci.
The LAMP assay presented here showed high sensitivity, and was a good tool to detect pneumococcal objectives. In a study by Seki et al., a LAMP assay targeting lytA was developed to detect S. pneumoniae, with acceptable results obtained at 60 minutes at 63°C, with a detection limit of 10 copies of S. pneumoniae DNA and sensitivity of 1000 times higher than that of conventional PCR (31). Regarding the Bsm polymerase used in the current study, the best temperature to produce loop structures was 62°C. In a study by Yang et al., a LAMP assay was developed in combination with a real-time technique targeting lytA, indicating acceptable reproducibility of the real-amp method (32).
The current study found that the reproducibility of the LAMP assay decreased when the reaction mixture was prepared under inappropriate conditions such as imperfect thawing of the dNTPs, MgSO4, and Bsm buffer solutions. Therefore, to avoid the user-associated errors, it is recommended that each of the solutions should be thoroughly thawed before adding into the reaction mixture. Wook et al., reported that the detection limit of the LAMP assay was significantly higher than that of PCR (33). Consistent with the results of Wook et al., the LAMP assay presented in the current study was also more sensitive than its PCR counterpart, and it was estimated that the detection of S. pneumoniae required a minimum of 5 copies of DNA. In the current study, the sensitivity of LAMP and PCR assays was 100%, and no DNA amplification was observed with non-pneumococci species, representing cbpM and lytA as specific to pneumococci and ideal for detection targets. The minimum period of time necessary to detect the turbidity of LAMP assay with the naked eye was 60 minutes, which was rapid compared with that of PCR. Indeed, this links back to the isothermal nature of LAMP, in as much as the polymerization of DNA proceeds at an optimum and constant temperature with no loss of time (34).
In conclusion, the robust performance of the LAMP assay in the current study suggested that sensitivity of the PCR and LAMP-based detection of S. pneumoniae was similar. However, the LAMP assay had a high detection limit, compared with the conventional PCR method.