1. Background
Staphylococcus aureus represents one of the most serious Gram-positive bacterial infections in nosocomial and community settings (1). The pathogenicity of S. aureus is caused by the expression of virulence factors, which can lead to superficial skin lesions or more-serious infections such as endocarditis, bacteremia, pneumonia, and osteomyelitis (2, 3). The development of high levels of penicillin resistance followed by the spread of strains resistant to the semi synthetic derivative of penicillin (e.g. methicillin) and other antibiotics has made the therapy of Staphylococcal infections an international challenge (4). Finding Methicillin resistant Staphylococcus aureus (MRSA) strains just two years after the clinical use of penicillinase-resistant Penicillin put specialists into difficulty with treating these infections (5). Vancomycin is considered the treatment of choice for MRSA cases, but recently there are reports on the emergence of vancomycin resistance in S. aureus isolates (6). The biofilm formation on host surfaces is considered an important virulence factor in S. aureus isolates (7, 8). Biofilm-forming Staphylococcus aureus strains are extremely difficult to eradicate because biofilm impairs antibiotic penetration and prevents normal immune responses (9, 10).
There are many methods to identify biofilm formation such as tube test, Congo Red Agar (CRA), Microtiter Plate test (Mtp), bioluminescent assay, and light or fluorescence microscopic examination (11, 12). These methods are often subject to severe analytical limitation and unable to detect bacterial adherence accurately (13, 14). However, the Mtp and CRA methods have been used to investigate biofilm formation more frequent than other methods (15-17).
In the present study, attempts were made to determine the prevalence of antimicrobial resistance and biofilm formation among MRSA isolated from children referring to a pediatric hospital in Tehran using Mtp and CRA methods.
2. Methods
2.1. Staphylococcus aureus Isolates
In total, 98 MRSA isolates were collected from 98 children referring to a pediatric hospital in Tehran during 2014 - 2015. All strains were examined by techniques appropriate for diagnosis of Staphylococcus spp. such as microscopic morphology examination, catalase test, coagulase test, mannitol fermentation on Mannitol Salt Agar and deoxyribonuclease (DNase) test (Merck, Germany) (18). All the isolates were confirmed to be S. aureus and MRSA by using PCR amplification of nuc and mecA genes, respectively (19, 20).
2.2. Antibiotic Susceptibility Test
Antimicrobial Susceptibility Test (AST) was performed according to the guidelines of clinical and laboratory standards institute (CLSI) (2013) (21). The used antibiotic disks in AST included penicillin (10 IU), cefoxitin (30 μg), clindamycin (2 μg), Linezolid (30 μg), minocycline (30 μg), trimethoprim-sulfamethoxazole (25 μg), gentamicin (10 μg), erythromycin (15 μg), and rifampicin (5 μg), all purchased from Rosco, Denmark. Vancomycin susceptibility test was performed by E-test (Liofilchem, Italy) and the results were interpreted according to the CLSI guidelines (2013). S. aureus ATCC 25923 was used as quality control.
2.3. Bacterial DNA Extraction
Genomic DNA was extracted from pure cultures using High Pure PCR Template Preparation Kit (Roche, Germany), according to the manufacturer’s guidelines.
2.4. Detection of nuc and mecA Genes
In the present study, all MRSA isolates were screened for the nuc and mecA genes using PCR and primers described in Table 1. A 25 μL PCR reaction mixture consisted of 12.5 μL of Master Mix (SinaClon, Iran), 2 μL of the DNA template, 1 μL of each primer (20 pmol), and 8.5 μL of ddH2O. DNA amplification was performed in a Thermocycler (Eppendorf, Hamburg, Germany) with an initial denaturation step at 95°C for 5 minutes, 30 amplification cycles each with 45 seconds at 95°C, 45 seconds at 54°C and 55°C for nuc and mecA genes, respectively, and 45 seconds at 72°C, followed by an additional extension step of 5 minutes at 72°C. The amplified products were electrophoresed on 1.5% agarose gel.
The gel was stained with Ethidium bromide (0.5 g/mL), visualized using UV transilluminator. The PCR products of nuc and mecA genes were sequenced and documented in Gen Bank, NCBI.
2.5. Detection of Biofilm Formation
2.5.1. Microtiter Plate Assay (Mtp)
The 98 clinical isolates of MRSA were screened for their ability to form biofilm using Mtp method according to Christensen et al. (1985) with some modifications (22). Briefly, bacterial isolates were grown in brain heart infusion (BHI) with 1% glucose (Merck, Germany) and incubated at 37°C overnight (13). Cultures were diluted 1:20 in fresh BHI-0.1% glucose. Then, 200 μL of the diluted solution was added to wells of a flat-bottomed polystyrene microtitre plate and incubated for 48 hours at 37°C. The negative control wells contained 200 μL of BHI-0.1% glucose. The wells were gently washed 3 times with phosphate-buffered saline (PBS; pH 7.2), fixed with sodium acetate (2%) for 10 minutes, dried at room temperature, and then strained with 0.1% crystal violet. After removing the crystal violet solution, the wells were washed with PBS to remove unbound dye. The optical densities (ODs) of the plates were observed at 630 nm using a Microtiter plate reader. Each assay was performed in duplicate. As a negative control, brain heart infusion broth with 1% glucose medium was used to determine the background OD. OD cut-off was then determined as an average OD of negative control + 3 × standard deviation of negative control. The OD cut-off value was separately calculated for each Microtiter plate. Biofilm formation by isolates was calculated and categorized according to the absorbance of the crystal violet-stained attached cells (23, 24) (Table 2).
| Cut-Off Value Calculation | Mean of OD Values | Biofilm Formation Abilities |
|---|---|---|
| OD > 4 × ODc | OD > 0.2 | Strong |
| 2 × ODc < OD ≤ 4 × ODc | 0.1 < OD ≤ 0.2 | Moderate |
| ODc < OD ≤ 2 × ODc | 0.05 < OD ≤ 0.1 | Weak |
| OD ≤ ODc | OD ≤ 0.05 | Negative |
2.5.2. Congo Red Agar Method (CRA)
Phenotypic production of biofilm in all MRSA isolates was assessed using culture on CRA plates as previously explained (12). Briefly, CRA plates were prepared by adding 0.8 g of Congo red (Sigma, USA) to 1 liter of brain heart infusion agar (Merck, Germany). Next, the CRA was autoclaved in 120°C for 15 minutes and then, 36 g of Saccharose (Merck, USA) was added using filtering. The plates were incubated for 48 hours at 37°C. The morphology of colonies was then interpreted based on colony color as pink, brown, and black (25, 26).
2.6. Statistical Analysis
The relationship between biofilm formation and drug resistance among MRSA isolates was evaluated by the Pearson Chi-Square test using SPSS version 21. P values less than 0.05 were considered significant.
3. Results
3.1. Samples Collection
In total, 98 clinical specimens were collected from blood (10.2%), throat (52%), wound (5.1%), sputum (5.1%), and other sources (27.6%). Participants were 52% girls and 48% boys. The mean age was 54 months. The youngest and the oldest patients had 1 month and 17 years of age, respectively.
3.2. Antibiogram Profile of Isolates
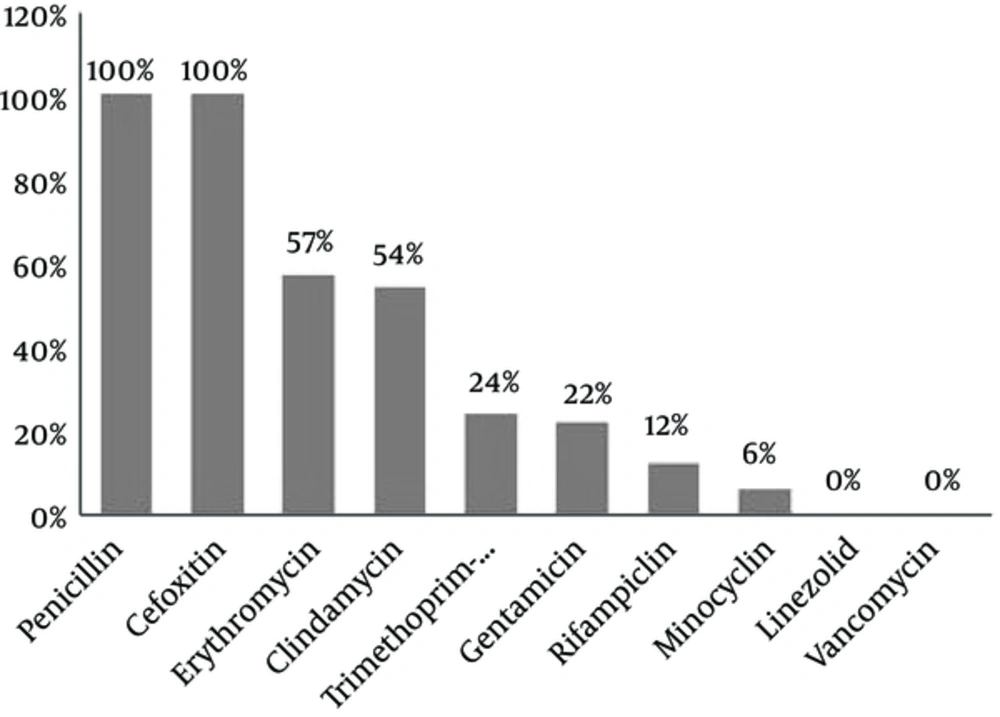
All the 98 (100%) MRSA isolates were found to be resistant to cefoxitin (30 μg) using disc diffusion method. Figure 1 shows the results of antibiotic resistance testing of the MRSA isolates for the nine antibiotics studied. The pattern of Minimal Inhibitory Concentration of vancomycin on the MRSA isolates was determined with concentrations varying from 0.19 to 2.5 μg/mL.
3.3. Detection of nuc and mecA using PCR
PCR revealed the presence of nuc and mecA genes (270 and 310 bp, respectively) in all 98 isolates (100%). The PCR products of nuc and mecA genes were sequenced and they were documented with Accession numbers KU163299 and KX024711, respectively, in Gen Bank, NCBI.
3.4. Detection of Biofilm Formation
3.4.1. Microtiter Plate Assay
The Mtp assay results showed that MRSA isolates were attached at different amounts. Attachment abilities were strong in 1 (1%) strain, moderate in 8 (8.2%) strains, and weak in 53 (54.1%) strains, while 36 (36.7%) of them had no attachment ability (Table 3).
| Method | Biofilm formation | Number | Percentage |
|---|---|---|---|
| Congo red agar method | Black | 56 | 57.1 |
| Brown | 24 | 24.5 | |
| Pink | 17 | 17.3 | |
| Microtiter plate assay | Strong | 1 | 1 |
| Moderate | 8 | 8.2 | |
| Weak | 53 | 54.1 | |
| Negative | 36 | 36.7 |
3.4.2. Congo Red Method
In this method, 57.1%, 24.5%, and 17.3% of MRSA isolates, respectively, showed black colonies (Strong biofilm producers), brown colonies (Weak biofilm producers), and red colonies (non-biofilm producers).
4. Discussion
The attachment and biofilm formation on surfaces causes bacterial resistance to inappropriate conditions such as antibiotics and immune response (27, 28). All of the 98 MRSA isolates were examined for their potential for biofilm formation. Using the Mtp method, 62 (63.3%) isolates were found to be biofilm producers among which, 9 (9.2%) strains were highly biofilm-positive (OD630 > 0.1) and 53 (54.1%) were low-grade biofilm producer, while 36 (36.7%) strains produced no biofilm. Other studies have reported a slightly higher number of biofilm producers among Staphylococcal species (23, 29).
In the CRA method, biofilm formation was observed in 80 (81.6%) isolates whereas 17 (17.3%) strains did not show any biofilm formation. This finding is in agreement with that of the study by Turkyilmaz et al. who detected biofilm production in 74.4% of isolates using CRA method (30). Among the phenotypic methods, Mtp assay has been reported as the gold standard for biofilm formation (14). CRA method is easy to perform and interpret, but due to its low specificity and sensitivity, we do not recommend it for detection of biofilm formation (31). When S. aureus assumes the biofilm phenotype, the associated infections are often extremely difficult to treat (32).
In the present study, antimicrobial resistance was 100% for penicillin and cefoxitin, 57% for erythromycin, 54% for clindamycin, 24% for trimethoprim- sulfamethoxazole, 22% for gentamycin, 12% for rifampicin, and 6% for minocycline, but all these isolates were susceptible to vancomycin and linezolid. Therefore, vancomycin, linezolid, and other glycopeptide drugs have remained the last resorts for treatment of S. aureus, especially MRSA-induced infections (33). A similar study was performed by Yousefi and his colleagues in 2016 in Iran. They determined the biofilm formation and antibiotic resistance pattern of Staphylococcus aureus isolated from urinary tract infection. They reported that 69.2% of S. aureus isolates were biofilm producers and resistance to four antibiotics, namely nitrofurantoin (71.4%/28.6%), tetracycline (57.7%/42.3%), erythromycin, and ciprofloxacin (56%/44) was higher among biofilm producers than among non-biofilm producers (29). These findings were consistent with our findings in the present study.
CharanKaur and Khare conducted a similar study in 2013 in India on biofilm investigation and antimicrobial susceptibility of MRSA isolates. Out of 231 isolates, 182 (78.8%) of the Methicillin resistant Staphylococcus aureus isolates were found to form biofilm (34).
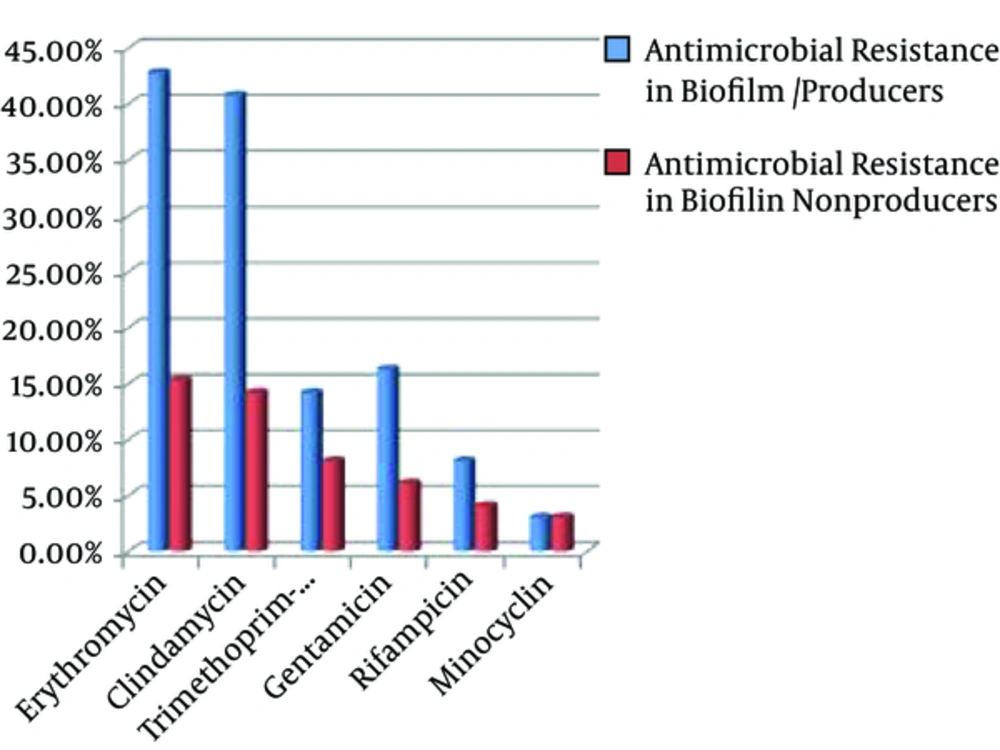
In our study, the antibiotic resistance pattern was higher in biofilm-producing MRSA than in non-biofilm-producers (Figure 2) although statistical analysis showed no significant relationship between biofilm formation and some antibiotic resistance; a finding that was previously reported by other researchers, too (9, 35).
The increased resistance of biofilm producing strains to antibiotics may be because the biofilm bacteria exhibit a slow rate of metabolism and divide infrequently, resulting in the decreased sensitivity to antibiotics targeted at cell wall synthesis (36). However, even antibiotics that target cellular functions, such as protein and DNA synthesis, cannot be effective on biofilm-producing organisms as they are at a quiescent state (37).
4.1. Conclusion
Unfortunately, the prevalence of Methicillin resistant Staphylococcus aureus isolates is rising and biofilm formation has an important role in the development of antibiotic resistance. Thus, early identification of these isolates and detection of biofilm formation by Mtp method are the essential steps towards the prevention of most serious nosocomial infections.