1. Background
Escherichia coli (E. coli) is the most important and the most common bacteria causing urinary tract infections (UTIs) (1). Signs and symptoms of this infection include chills, fever, flank pain, and dysuria (2). For acute respiratory, enteric, and urinary tract infections as well as serious systemic infections, such as bacteremia, fluoroquinolones have been identified as the first and second line of antibacterial treatment (3). The bacteriostatic effect of quinolones, is the result of their role in trapping of DNA gyrase to form reversible drug-enzyme-DNA cleavage complexes. In a continuous process, cell death arises from chromosome fragmentation in protein synthesis (dependent or independent pathways), according to the distinguished quinolone structures. In the former pathway, reactive oxygen species kill bacteria because of irreversible oxidative DNA damage. As an additional lethal action, mazEF (toxin–antitoxin) is triggered. The save our souls (SOS) responses and other stress responses, support the bacterial survival and resistance development (4). Resistance associated mutations (RAMs) in the case of fluoroquinolones can be divided roughly to two parts: A) E. coli resistance to fluoroquinolones is caused mainly by chromosomal mutations (5). An essential enzyme for DNA replication in E. coli is DNA gyrase (DNA topoisomerase II), which is an ATP-dependent enzyme that induces negative supercoils to separate the two daughter duplexes (6). Both subunits of DNA gyrase (subunit A and B) are essential for cell viability and double strands separation in DNA replication and transcription in E. coli. Also, they poses β-hemolytic activity (ability to lyses blood cells in blood agar) and thus may contribute to the virulence properties of isolates (7, 8). Most frequently, ciprofloxacin resistant E. coli isolates carry mutations in gyrA; this is located in a region, which is called the quinolone resistance determining region (QRDR) (9, 10). B) Plasmid-mediated quinolone resistance gene, named qnr, was reported in 1998. This gene encodes a 218-amino acid protein, which was later renamed QnrA; it is a member of the the penta peptide-repeat family. More recently, four additional proteins, QnrB, QnrS, QnrC, and QnrD, have been identified in several Enterobacterial species (11-15). These proteins interact with quinolones, topoisomerases, and DNA, and act by limiting the binding of quinolones to their targets (16). By itself, the qnr gene confers low-level resistance to quinolones. However, the presence of this gene facilitates the acquisition of high-level resistance among initially susceptible strains (17, 18).
Other mechanisms include multidrug resistance efflux pumps OqxAB and QepA (19, 20) and acetylation of the piperazinyl amine of ciprofloxacin and norfloxacin by a new variant AAC (6’)-Ib-cr of the aminoglycoside acetyltransferase (21). Because of the key role of updated information of resistant rate and related resistant genes in treatment and prevention policy, this study aimed at evaluating the contribution of gyrA and qnrA genes among ciprofloxacin resistant E. coli isolates from patients with UTIs of Imam Khomeini hospital of Tehran.
2. Methods
2.1. Sample Size
The sample size was calculated, based on the frequency of E. coli isolates for UTIs, using the following formula:
N = (1.96) 2 (0.7) (0.3) / (0.1) 2 = 80.6 ~ 80 (E. coli isolates)
2.2. Sample Collection
This descriptive study was done during September to March 2014, and 80 E. coli strains were isolated from 100 midstream urine samples of out/inpatients that referred to Imam Khomeini (IK) hospital. Based on age, all patients were identified as “adults” when they were over 18 years old and “children”, if they were under 17.9 years old.
All urine samples from outpatient and in-patient wards were cultured on MacConkey agar (Merck Company). Escherichia coli identification was done by using standard tests such as: oxidase test, TSI agar (Merck Company) cultivation, urease test, motility, and IMViC tests (22).
All E. coli isolates were transferred to cryovials, containing Trypticase soy broth (TSB) plus 15% glycerol and kept at -70°C, for further evaluation.
2.3. Antibiotic Susceptibility Test (AST)
The resistance/sensitivity of the isolates to 20 common antibiotics was investigated by standard disk diffusion technique (Kirby–Bauer method), according to clinical and laboratory standards institute (CLSI) 2014 guidelines. All disks were purchased from Merck Company (England). The antibiotics used for antimicrobial susceptibility test (AST) included ceftriaxone (30 µg), piperacillin (100 µg), gentamicin (10 µg), ciprofloxacin (5 µg), ampicillin (10 µg), amoxicillin-clavulanic (20/10 µg), cefazolin (30 µg), aztreonam (30 µg), cefepime (30 µg), trimethoprim/sulfamethoxazole (1.25/23.75 µg), nitrofurantoin (300 µg), chloramphenicol (30 µg), imipenem (10 µg), meropenem (10 µg), ertapenem (10 µg), norfloxacin (10 µg), tetracycline (30 µg), and cefixime (5 µg). All antibiotic disks were purchased from Mast Company, UK. Suspension of each bacterial isolate was prepared with a turbidity equal to 0.5 McFarland standard (1.5×108 CFU/mL), and next, a lawn of bacteria was cultured on a Mueller–Hinton agar (Merck Company) by a sterile cotton swab. By a sterile forceps, selected antibiotic disks were sited on the agar media. All plates were incubated at 37°C for 18 to 24 hours. After that, the diameter of the inhibition zone around each disk was measured and the results were compared to the standard CLSI criteria and reported as sensitive (S), intermediate (I), or resistant (R) (23). Escherichia coli ATCC 25922 was used as a quality control strain.
2.4. Minimum Inhibitory Concentration (MIC) of Ciprofloxacin by E- test Method
The minimum inhibitory concentration (MIC) of ciprofloxacin was determined for ciprofloxacin– intermediate and resistant isolates in AST by the E-test strip method (Liofiche, Italy). The E-test procedure was done based on the manufacturer’s protocol. According to CLSI 2013 guidelines, MIC ≤ 1μg/mL was considered as sensitive, ≥ 4 μg/mL as resistant, and equal to 2 μg/mL as intermediate (23). As a quality control, E. coli ATCC 25922 was used again.
2.5. DNA Extraction, Polymerase Chain Reaction (PCR) Technique and Sequencing
Bacterial chromosomal DNA bacteria was extracted using the boiling method for further PCR of the gyrA gene (24). The Gene Jet Plasmid (Miniprep ≠ K0502 kit, Thermo scientific, Lithuania) was used to extract plasmid for further PCR of the qnrA gene. The Mastermix (PR901638/reaction 200, SinaClon, Iran) was used for the PCR procedure. For quality control, E. coli ATCC 25922 was used, simultaneously. The list of primers, PCR programs, and the PCR products are mentioned in Tables 1 and 2. The PCR products of the studied genes were determined after electrophoresis on 1% agarose gel and visualized under UV radiation of gel documentation. Further sequencing was done by the Bioneer Company (Korea). Further nucleotide analyzes were done with the Chromas 1.45 software and BLAST in NCBI. After that, genes submission was done in NCBI.
| Step | Temperature (°C) | Time (min) |
|---|---|---|
| Primary denaturation | 94 | 3 |
| Denaturation | 94 | 1 |
| Primers coupling | 58 | 1 |
| Polymerization | 72 | 1 |
| Final polymerization | 72 | 5 |
2.6. Statistical Analysis
This research was a descriptive study. Results were presented as frequency (percentage) for categorical variables with 95% confidence intervals (95% CI). For the statistical analysis, the statistical software SPSS version 16.0 (SPSS Inc., Chicago, IL) was used. The P values of 0.05 or less were considered statistically significant.
3. Results
The prevalence of UTIs by gender was as follow: 51% in females, 28% in male, and 21% in children. Urine samples were collected from different hospital wards as follow: 35% of females in the general ward, 15% urology, 15% neurology, 10% other sections, and 25% outpatient. Furthermore, 79% of patients were adults and 21% were children.
The results of each test were as follows:
3.1. Phenotype Confirmation
Of 100 urine samples, 80 E. coli isolates were detected and identified. The E. coli identification confirmation was done based on lactose positive colonies on MacConkey agar, TSI A/A gas + H2S-, indole and MR and motility positive, VP, and citrate negative (22).
3.2. Antimicrobial Sensitivity Test (AST)
Briefly, the highest resistance were detected against ampicillin and piperacillin (85.3% and 85%, respectively). All isolates were sensitive to three tested carbapenems (100%) and 98.7% to nitrofurantoin, as a usual drug in treatment of UTIs. Around 80% of isolates were resistant to three antibiotic families and were identified as multi drug resistant (MDR). The AST results for all tested antibiotics were shown in Table 3.
| Antibiotics | Susceptibility | Intermediate | Resistance |
|---|---|---|---|
| Ertapenem | 100 | 0 | 0 |
| Meropenem | 100 | 0 | 0 |
| Imipenem | 100 | 0 | 0 |
| Nitrofurantoin | 98.7 | 0 | 1.3 |
| Amikacin | 83.5 | 5.1 | 11.4 |
| Chloramphenicol | 72.5 | 5 | 22.5 |
| Cefepime | 36.2 | 21.3 | 42.5 |
| Ofloxacin | 32.6 | 0 | 67.4 |
| Gentamicin | 35 | 2.5 | 62.5 |
| Aztronam | 27.5 | 3.8 | 68.7 |
| Ceftriaxone | 26.2 | 1.3 | 72.5 |
| Tetracycline | 25 | 0 | 75 |
| Cefazoline | 23.8 | 2.5 | 73.7 |
| Trimethoprim/sulfamethosazol(SXT) | 21.3 | 0 | 78.7 |
| Ciprofloxacin | 18.7 | 3.8 | 77.5 |
| Piperacillin | 12.5 | 2.5 | 85 |
| Ampicillin | 10 | 6.2 | 83.8 |
aValues are presented as %.
3.3. Minimum Inhibitory Concentration Determination by the E-test
The MIC of ciprofloxacin ranged from 0.002 μg/mL to 32 μg/mL. The average minimum inhibitory concentration was 0.44 ± 0.93 with scope of 0.008 to 4.0. This study found that 61.76% of tested isolates were resistant to ciprofloxacin and showed MIC of ≥ 4 μg/mL.
3.4. Assessment of gyrA Gene
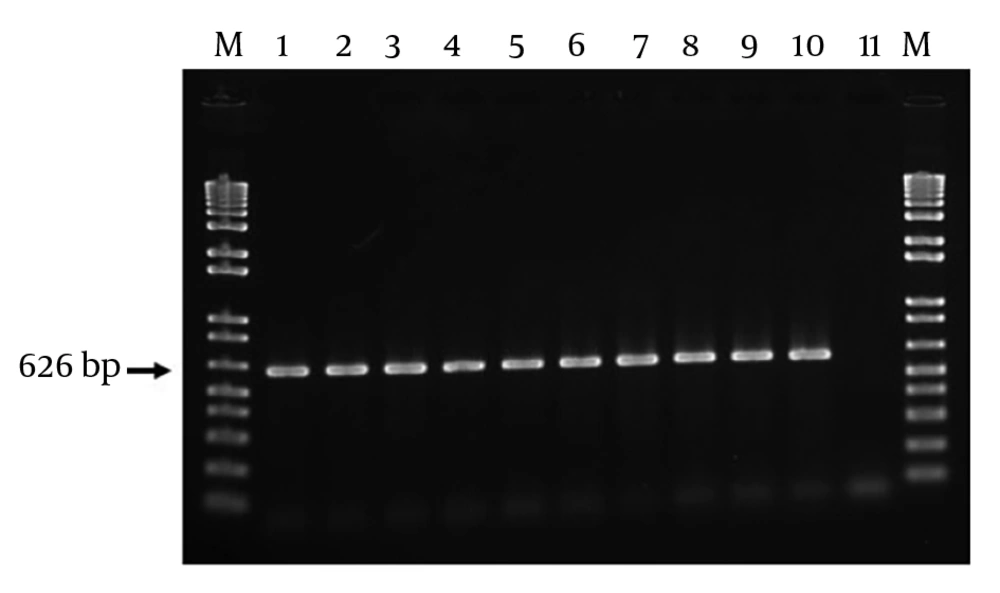
Of 80 E. coli isolates in this study, a 626-bp band was detected among 100 of isolates, which confirmed the existence of intact gyrA gene.
3.5. Prevalence of qnrA Gene
Only 39% (31/80) were confirmed to have qnrA gene after detection of a 516-bp band. The prevalence of the qnrA gene is shown in Figure 2.
3.6. Blasting and Gene Submission
Sequencing analysis was done by Chromas 1.45 software and further BLAST at NCBI. It was clear that some mutation was seen, yet the mean full change was related to change of serine to leucine at position 83 (Ser- 83 to Leu). The sequence of samples was 100 similar to the sequence of E. coli with accession number GQ869684. The sequence of this study was registered in NCBI as GenBank: KX587467.1. For more information, please refer to www.ncbi.nlm.nih.gov/nuccore/KX587467.
4. Discussion
In this descriptive study, by an antimicrobial sensitivity test (AST) and microdilution test, the antibacterial pattern, and by further PCR method, the role of gyrA and qnrA genes among 80 E. coli isolates from patients of the Imam Khomeini hospital of Tehran were evaluated. Based on the AST results, the highest resistance was towards ampicillin and piperacillin (85%). Carbapenems were the most effective antibiotics (100% susceptible). The gyrA and qnrA gene were detected in 100% and 39% of ciprofloxacin resistant E. coli isolates, respectively.
A study at Mansura hospital, in Egypt, was done in order to determine the pattern of antimicrobial resistance to common antibiotics and to verify the mechanism of resistance. In that study, the main mechanism of resistance to quinolones was related to the mutation in the chromosomal quinolone resistant determinant region (QRDR). Moreover, there was high resistance because of plasmid gene’s transformation (10). Similar to a recent study, both mutated gyrA genes accompanied by qnrA genes were responsible in ciprofloxacin resistance among E. coli isolates.
Winissorn et al. verified the presence of qnrA gene and its relationship with intl1 in resistant isolates of E. coli. Also, simultaneous existence of qnrA gene and ESBL-producing genes were detected. However, the frequency of qnrA gene among E. coli isolates was 8 (25). In the current study, neither intl1 nor the ability of ESBL generation was investigated, yet, despite Winissorn’s results, the frequency of qnrA gene among E. coli isolates in a recent study was higher (39% versus 8%). This may be related to the time or geographic difference and the variation of antibiotic prescription pattern among the two countries.
In the study of Kariuki et al. from Kenia, Uropathogenic E. coli (UPEC) isolates from patients, with urinary tract infections (UTIs), were evaluated by phenotypic AST and molecular PCR and PFGE methods. Bacteria, which had double mutation in their two amino acids (replacement of serine to leucine and aspartic acid to asparagine) of QRDR domain of chromosomic gyrA gene, were identified as quinolone resistant bacteria (26). Despite Kariuki et al.’s study, only one of the two mentioned mutation types, conversion of serine to leucine at position 83, was seen in a recent study.
From 2002 to 2004 in Minnesota of USA, 931 Escherichia coli isolates were collected from two groups of patients and control, consequently the resistance rate against common antibiotics was investigated. The highest resistant rate was against fluoroquinolones, trimethoprim/sulfamethoxazole and cephalosporins, specifically among people, who were hospitalized (27). Similar to the recent study, of 80 E. coli isolates from 100 patients with UTIs, who referred to Imam Khomeini hospital of Tehran, resistance to ciprofloxacin (77.5%) was the third highest among E. coli isolates of the recent study.
In year 2010 to 2011, a study on 200 patients with urinary tract infections was conducted in Gorgan, Iran, including Escherichia coli isolates from UTI cases; resistance to Nalidixic acid, nitrofurantoin, clindamycin, tetracycline, cotrimoxazole, cefotaxime, cefazoline, ceftazidime, ceftriaxone, ciprofloxacin, and cefepime was 61%, 1.3%, 98.7%, 65%, 61%, 30%, 38.5%, 23.2%, 27%, 38.5%, and 23.3%, respectively. All samples were susceptible to imipenem (27).
In this study, although the level of resistance to all of the tested disks was not similar to a research from Gorgan, yet the resistance percentage against some similar tested antibiotics, such as sulfamethoxazole trimethoprim (78.7%), cefazolin (73.7%), ceftriaxone (72.5%), ciprofloxacin (77.5%), cefepime (42.5%) was higher. Also, in both studies, all the E. coli isolates were sensitive to imipenem.
The obtained results from this study showed higher percentage of resistance against the same antibiotics rather than Gorgan’s study and this difference may be related to the time difference as there was about four years difference between these researches.
Also, another reason for this variation could be related to the length of use of antibiotics, such as quinolone, along this time, which could have caused the emergence of new resistant strains against these drugs, especially ciprofloxacin.
Fortunately, despite of the increasing resistance rate to some antibiotics, this group of bacteria are sensitive to imipenem.
Warburg et al. (1999 to 2005), verified that resistance of E. coli strains against fluoroquinolones was related to the aac (6’)-lb-cr , qnrA, and qnrB genes by the PCR method. Also, they showed that simultaneous fluoroquinolones resistance and the ability to produce ESBLs is going to increase. As the mentioned genes are settled on the plasmid, transformation of plasmid genes plays an important role in this process. It even seems that synchronous resistance against fluoroquinolones and the ability to produce ESBLs were because of the relationship of the two plasmids (28).
In the study of Alheib, the qnrB gene was present in 83.83% of E. coli strains, no qnrA and qep A were detected, and aac (6’) Ib was the most common plasmid gene of quinolone resistance (29).
However, in a recent study, the simultaneous existence of aac (6’)-Ib-cr and qnrA gene was not verified yet in another research done by “Hakemi et al. (not published yet), the frequencies of qnrA gene and aac (6’)-lb-cr genes were 39% and 72%, respectively.
In the study of Cremet et al., co-existence of plasmidic quinolones resistant gene among E. coli isolates with ESBL related genes with low sensitivity to fluoroquinolones were verified. Although they did not report anything about the frequency of qnrA or qnrB genes, they reported that most cases of resistance was due to the presence of plasmidic aac (6’) –lb gene (30).
In this study, the frequency of qnrA gene was 39% and no qnrB was detected; such variation may be because of difference of resistance mechanism between E. coli isolates in Iran versus Kuwait and Syria. However, difference of time and antibiotics consumption pattern must be included. In addition, such variations may be related to the origin of bacterial isolation other than urine.
Also, a similar mutation style at position 83 of gyrA gene (serine 83 lucine) was reported by Crement et al. in France, similar to the mutation type in a recent study (31).
Liu et al. evaluated mutations in gyrA and parC genes of E. coli isolates and showed that the number of mutations in gyrA and/or parC was significantly associated with MIC of quinolones (31).
Similarly, in a research by Cattoir et al. at Pasteur institute of France, three isolates of 64 isolated Enterobacter (another member of the Enterobacteriaceae family), which were isolated from Kuwait during a shared study, were positive for qnrB gene (4.7%) and no qnrA gene was detected (32).
Also, Dashti et al. from Edinburgh showed that 69 Klebsiella pneumoniae isolates were detected from different clinical samples and their ESBL-producing ability were verified. Furthermore, 32 isolates were resistant to ciprofloxacin and existence of double mutations in two positions of 83 and 87 were identified as the cause of ciprofloxacin resistance (33). However, the bacterial strains were different, yet both were members of Enterobacteriaceae and showed shared mutation of the gyrA gene at position 83.
Pakzad et al. verified the distribution of plasmid genes “qnr” and their increase among E. coil quinolone resistance. They showed that qnrA gene had a more significant role to provide resistance against quinolones rather than qnrB (34). Similar to Pakzad’s study, the only plasmid gene responsible for ciprofloxacin resistance among E. coli isolates in the current study was qnrA. Although determination of the frequency of qnrB gene was not an aim of this study yet it was not detected (0%) in the author’s previous study among E. coli isolates from UTIs (35).
Firoozeh et al. from “Khorramabad”, Iran, showed that 45% of 140 E. coli isolates were resistant to ciprofloxacin versus 77.5% in a recent study. Moreover, 116 of 140 E. coli isolates were resistant to nalidixic acid, yet not to ciprofloxacin. Also, gene frequencies of qnrA and qnrB were 14% and 9%, respectively. However, in the current study and studies by Abdi et al. and Firoozeh et al., there was a high difference between the frequency of rate of resistance to ciprofloxacin and the frequency of the qnrA gene (39% versus 14%) (1, 36). It seems that geographic differences even in the same country, time differences and also, the difference between using disks and experiment methods may influence the results and must be included in the interpretation of such variations.
Based on a study by Pourahmad, from Sharkord, three types of mutations were detected in the gyrA gene, including Ser-83 to Leu, Tyr-50 to Phe, and Ala-119 to Glu (37). All of the mentioned mutations were detected in Quinolone Resistance Determining Region (QRDR), which is very close to GyrA active site (38). The most frequent mutation type was Ser-83 to Leu.
In the study of Fu et al., different patterns of mutations of gyrA were identified, yet only some of these mutations were involved in fluoroquinolons resistance. Two single mutations of the Ser- 83 to Tyr and Ser-83 to Leu were related to ciprofloxacin resistance (38). Detection of Ser-83 to Tyr in QRDR of E. coli isolates in the recent study was in accordance with Pourahmad and Fu et al.’s studies.
Mutations may influence the molecule, however, some mutations are uneffective. A kind of mutation (Ser-83 to Leu), which was detected in the gyrA gene, which includes a change from a polar to a non-polar acid amine, may contribute to GyrA function; the isolates, which were candidates for sequencing had MIC of ≥ 4μg/mL for ciprofloxacin.
In conclusion, contribution of both mutated chromosomal gyrA genes and plasmidic qnrA resistance genes in some of the high ciprofloxacin resistant bacterial strains in this study besides the overuse of antibiotics, can increase the emergence of resistant strains.