1. Background
Cystic fibrosis (CF) is a complex genetic disorder with pulmonary involvement. CF patients have normal lungs at birth; but recurrent and chronic bacterial respiratory infections lead to progressive respiratory compromise, long-term morbidity, and mortality in the patients (1, 2). Several bacterial species can colonize the respiratory tract of CF patients. The testing of respiratory secretions from these patients is a suitable method for detecting colonization with opportunistic bacteria (2). Pseudomonas aeruginosa is the main respiratory pathogen although other Gram-negative bacteria such as Burkholderia cepacia, Stenotrophomonas maltophilia, and Acinetobacter baumannii are also detected in the respiratory tract and play key roles in pulmonary pathology (1-3). P. aeruginosa can persist for a long time as a chronic infection to impair lung function (3, 4). This microorganism is acquired from the environment or through patient-to-patient transmission (5). Rarely may Acinetobacter spp. be found in the respiratory secretions of CF patients (6). The genus Burkholderia is an adaptable Gram-negative bacterium, with some species such as Burkholderia cepacia causing life-threatening infections in CF patients. Infections caused by these bacteria can be difficult to treat because of their significant antibiotic resistance (6, 7). Although the prevalence of colonization with Burkholderia cepacia complex is relatively low in CF patients, it is important as some genomovars of this bacterium can reduce survival after lung transplantation (8). Stenotrophomonas maltophilia is another bacterial species that commonly colonizes the respiratory tract of CF patients. However, the effect of this opportunistic pathogen on the development of CF lung disease has not yet been completely specified (9).
Rapid detection and identification of pathogenic microorganisms in CF patients not only help prevent the patient-to-patient transmission, but also expedite the prompt institution of appropriate treatment for infected patients. Late detection impedes the eradication of these pathogens and leads to chronic infection (5). Culture-based methods are time-consuming in the detection of bacteria from the airways of CF patients and they usually give false positive and false negative results due to colonization and previous antibiotic use, respectively. This is while molecular techniques can detect causal bacteria rapidly (10, 11). Polymerase chain reaction (PCR) assays targeting the 16srRNA gene can be used for the detection of lung infection bacteria in CF patients (10, 12).
2. Objectives
This study aimed to utilize a molecular detection technique for the identification of Gram-negative bacterial flora (P. aeruginosa, B. cepacia complex, Stenotrophomonas maltophilia, and Acinetobacter baumannii) in the airways of CF patients.
3. Methods
3.1. Study Population and Bacterial Isolation
This cross-sectional study was conducted on 64 CF patients aged 4 months to 23 years referring to Mofid Children’s Hospital in Tehran as outpatients or CF patients hospitalized from September 2013 to March 2014. We examined sputum, deep pharyngeal swabs, and Bronchoalveolar lavage (BAL) samples of these patients.
The samples were immediately transferred to the Pediatric Infections Research Center (PIRC) laboratory in BHI medium cryotube in a specific box, and then conserved in a freezer at a temperature of -80°C.
3.2. DNA Extraction
The DNA extraction was performed using the Intron DNA extraction kit (Korea, cat. No. 17045) according to the manual protocol.
3.3. PCR Assays
First, the 16srRNA gene was detected in all samples to identify bacterial infection (12). The primers are shown in Table 1. Then, Pseudomonas spp., A. baumannii, B. cepacia, and S. maltophilia were identified by specific primers (Table 1). The PCR was performed for each sample in the following mixture: 1X Specific PCR buffer, 0.4 mM of the dNTPs mix, 0.7 mM of MgCl2, 1.6 M of each primer, one unit of Taq polymerase enzyme, 2 µL of DNA, and sterile distilled water to the final volume of 25 µL. The PCR condition was as follows: initial denaturation at 94°C for 5 minutes for 30 cycles consisting of denaturation (94°C for 45 seconds), annealing (48°C, 61°C, 53°C, 53°C, and 50°C for 45 seconds for the 16srRNA gene, A. baumannii, S. maltophilia, B. cepacia complex, and Pseudomonas spp., respectively), extension (72°C for 45 seconds), followed by a final extension at 72°C for 7 minutes. The internal positive control was used in the PCR assays.
| Primer | Sequence (5’→3’) | PCR Product Size (bp) | References |
|---|---|---|---|
| 16S rRNA-F | CCTCTCAGACCAGTTA | 250 | (13) |
| 16S rRNA-R | CCTAACACATGCAAGTCGA | ||
| Acinetobacter baumannii-F | TAATGCTTTGATCGGCCTTG | 353 | (14) |
| Acinetobacter baumannii-R | TGGATTGCACTTCATCTTGG | ||
| Pseudomonas spp.-F | GTCTTTCAGCTCGAT | 536 | (15) |
| Pseudomonas spp.-R | ATGAAGCTTCGTCCTCTGCAT | ||
| B. cepacia complex-F | AGRGTTYGATYMTGGCTCAG | 463 | (16) |
| B. cepacia complex-R | CCGRCTGTATTAGAGCCA | ||
| Stenotrophomonas maltophilia-F | TACCACCCGTACCTGGACTT | 149 | (16) |
| Stenotrophomonas maltophilia-R | ATCGCATCGTTGCTGTTGTA |
Finally, the PCR products were evaluated on a 1.5% agarose gel.
4. Results
In this cross-sectional study, 64 clinical specimens of sputum, deep pharyngeal swabs, and BAL were obtained from 64 CF patients. Some specimens were unsuitable for analysis due to inappropriate collection techniques that were excluded from the survey. Therefore, 51 specimens from 51 patients were examined. The mean age of the patients was 6.7 ± 5.2 years. There were 28 (55%) male patients in the sample. All samples were positive for 16srRNA; thus, bacterial infection was confirmed.
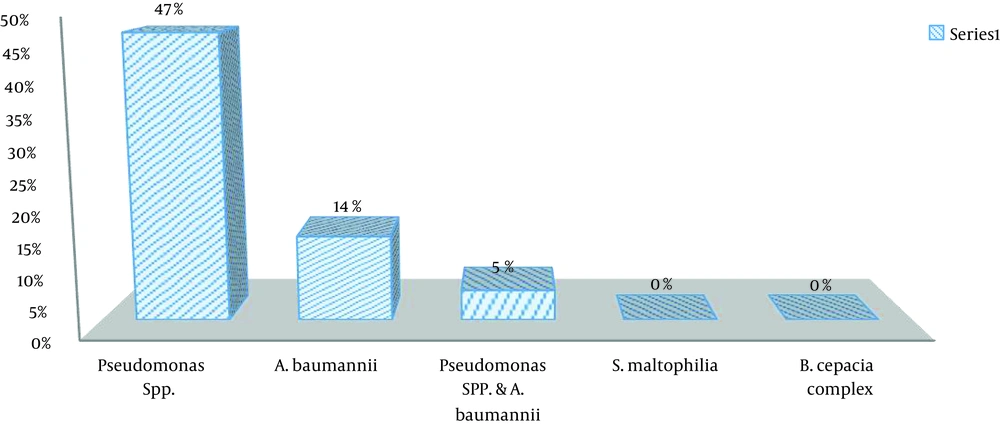
Pseudomonas spp. was detected in 30 (46.87%) samples and A. baumannii in nine (14%) samples (Figure 1). Co-colonization by Pseudomonas spp. and A. baumannii was observed in three (5%) samples. S. maltophilia and B. cepacia complex were not detected in any of the samples (Figure 2).
5. Discussion
The PCR assay is a rapid and accurate method for detecting bacterial infections in CF patients’ airways (17). Molecular tests are not accurate modalities to differentiate between infection (asymptomatic colonization) and disease; however, even the detection of colonization is essential in these patients. Our study aimed to determine the presence of Gram-negative bacteria in CF patients whether as colonization or as a disease. Previous studies utilized 16srRNA to confirm the presence of bacterial infection in respiratory tract specimens of CF patients (4, 18).
In our study, all samples were positive for 16srRNA, confirming the presence of bacteria in the respiratory tract of all patients. In the study by Bittar and Rolain in France, 96.8% of the samples were positive for bacterial infection in sputum samples of CF patients according to 16srRNA detection results (17). These results are similar to our survey and indicate the high prevalence of bacterial colonization in CF patients.
Based on the molecular investigation, P. aeruginosa was the most frequent species in the sputum of CF patients in our study. Keating et al. in the United States in 2016 reported that 38% of 554 children with CF were positive for P. aeruginosa (2). Cigana et al. in 2016 from Italy detected P. aeruginosa in 50% of 338 CF patients with a mean age 26.7 years (5). In 2011, a study from Germany reported a detection rate of 52% for Pseudomonas spp. in 165 children and adult patients (9). Khalilzadeh et al. in 2012 assessed 22 children and young adults and confirmed that 45.5% of the hospitalized CF patients had P. aeruginosa (19). The results of these three studies are comparable to the findings of the current study. In a study conducted in Tehran by Khanbabaee et al., P. aeruginosa was again found to be the most prevalent species isolated from 129 pediatric CF patients (20).
Contrary to the above studies, Forozesh Fard et al. in 2012 in Iran showed that only were 26% of 27 CF patients infected with P. aeruginosa (21). Moreover, a detection rate of 13% was reported for P. aeruginosa by Deschaght et al. in 2010 from Belgium in a study conducted on 397 adult and pediatric CF patients (22). The molecular results in our study showed a high rate of colonization with Pseudomonas spp. in CF patients. In a systematic review of 46 studies (1823 patients) from 16 countries in 2017, P. aeruginosa was one of the most commonly detected bacteria isolated from noses and sinuses of CF patients (23). This difference could be attributed to various factors, including differences in the frequency of Pseudomonas spp. in the patients’ living environment, age, the health status of patients at the time of the study, and the frequency of their contacts with other patients infected with Pseudomonas strains.
In the current study, 14% of the isolates were attributed to A. baumannii. The results of a study on U.S CF patients between 2004 and 2008 showed that 9% of isolates were A. baumannii-positive (24). A case report of multi-drug-resistant (MDR) A. baumannii isolated from a CF patient was published in Italy (25). The results of these studies point to a low prevalence of A. baumannii in CF patients.
B. cepacia was not detected in samples collected from our patients. The rate of colonization with B. cepacia has been reported low in other studies, as well (24). However, Eram et al. from Iran reported a prevalence of 12.25% for B. cepacia and 24.5% for P. aeruginosa in children with CF (26). Interestingly, B. cepacia was isolated from the sputum of a 14-year-old CF patient in Ireland in 2016 (21).
Zhao et al. published a systematic review on the co-colonization of P. aeruginosa and Aspergillus fumigatus in CF in 2018 (27). They concluded that the co-colonization of P. aeruginosa and A. fumigatus occurs in 15.8% of CF patients (27). The rate of co-colonization of P. aeruginosa and A. baumannii in our study was 5%. These Gram-negative bacteria can change into multidrug-resistant (MDR) or extensively drug-resistant (XDR) strains as a result of conferring antibiotic resistance genes, which makes a very serious therapeutic problem. There are major variations in the co-colonization rate in different studies according to the time of the study, but the rates may be similar in various geographical regions.
5.1. Conclusions
The results of the current study showed that according to molecular assay, Pseudomonas spp. were the most prevalent Gram-negative bacteria isolated from the sputum, deep pharyngeal swabs, and BAL samples of CF children. Early detection of colonization may prevent serious respiratory infections in children with CF.