1. Background
Toxoplasma gondii is a compulsory intracellular parasite with global spread that causes toxoplasmosis infectious disease in human beings and animals (1). Toxoplasmosis can occur by eating food and/or water contaminated with parasite oocytes, as well as the consumption of meat containing the parasite tissue cysts (2). Predominantly, there is no symptom in healthy individuals, yet the parasite can cause severe toxoplasmosis in people with immunodeficiency, such as those who are receiving chemotherapy, organ transplant recipients, patients with cancer, and patients infected with human immunodeficiency virus (HIV) (3, 4).
The rapid detection of toxoplasmosis, especially in patients with immunodeficiency is considered as an important approach in treatment of the infection (5). Routinely, the diagnosis of the infection is based on the measurement of antibodies against T. gondii in the serum by serology methods, such as immunofluorescence assay (IFA) (6) and enzyme-linked immunosorbent assay (ELISA) (7). Since T. gondii antigens and its specific antibodies may not be present early in toxoplasmosis, the serology methods had some problems and limitations (5) while interpreting the results of the methods was problematic in pregnant women (8).
During the last decade, molecular methods have progressed in the diagnosis of toxoplasmosis, particularly in congenital diseases; among these techniques, the nested-PCR method is one of the sensitive techniques for the detection of infection (9). There are different sequences for specific and sensitive diagnosis of toxoplasmosis. Previous studies demonstrated that the use of multi copy target was more sensitive than single-copy (9). It has been established that the RE sequence is repeated 200 to 300 times in the genome, while the sequence is more sensitive than the B1 sequence, therefore, it is more suitable for the diagnosis of toxoplasmosis (10). Based on the above explanations and the different genotypes of the parasite in Iran (11) and other countries (12, 13), the aim of this study was to evaluate the frequency and genotyping of T. gondii in blood and CSF samples of children with immunodeficiency, using the molecular and RFLP methods.
2. Methods
2.1. Ethical Aspects
All experimental procedures were confirmed by the Ethics Committee of Ahvaz Jundishapur University of Medical Sciences (with OG-93121 code). Informed consent was obtained from parents of the children involved in this study.
2.2. Study Population and Samples Collection
The current research was a descriptive cross-sectional study. A total of 170 blood samples and 170 cerebrospinal fluid (CSF) samples were randomly collected from 170 children with cancer while receiving chemotherapy at Shafa Hospital of Ahvaz (Southwest of Iran), during December 2015 to June 2016. The sampling was performed based on clustering probable sampling, and sample size was selected based on the estimated number of patients at the treatment center. After sampling, the collected samples were transferred to the Department of Parasitology of Ahvaz Jundishapur University of Medical Sciences. Five milliliters of blood samples containing anticoagulant material were centrifuged and their buffy coats were separated and 1 to 1.5 mL CSF samples were kept in specific vials. Finally, all samples were stored at -20°C.
2.3. DNA Extraction
DNA extraction was performed using the Bioneer kit (South Korea) through the following procedure: 100 µL of blood samples (buffy coat) and CSF were mixed with 20 µL of proteinase k. Then, 400 µL of buffer solution was added and placed at 60°C for 10 minutes. The samples were centrifuged at 12000 rpm for five minutes. Afterwards, the supernatant was removed and 400 µL of binding buffer solution was added and placed at 60°C for 10 minutes. In the next stage, 100 µL of isopropanol was added and centrifuged at 8000 rpm for one minute. The solution was poured in the column separator and 500 µL of washing solution was added (w1) and centrifuged at 8000 rpm for one minute. Then, 500 µL of washing solution (w2) was added and centrifuged at 8000 rpm for one minute. Finally, 50 µL of elution buffer was poured off and centrifuged at 8000 rpm.
2.4. Polymerase Chain Reaction
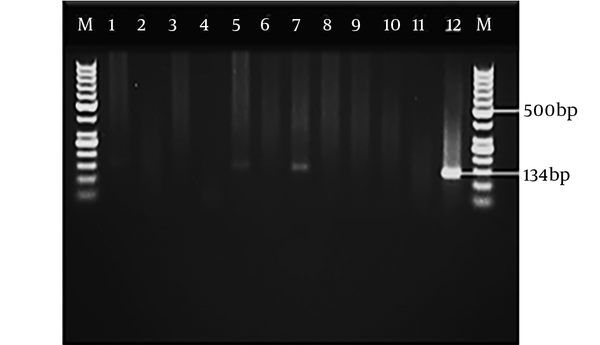
The PCR assay was carried out in a final volume of 20 µL, containing the master mix (Accu Power Lyophilized Premix of Bioneer Company, Korea), Taq DNA Pol 1.0 unit, 10 mM Tris-HCl (PH: 9.0), 30 mM KCl, 1.5 mM MgCl2, 1 µM each primer (Fermentas Company, Germany), 250 µM dNTPS, and 100 µg/µL of extracted DNA. The PCR assay was performed based on the RE sequence, using primer pairs including (TOXO F2 5’ AGG CGA GGG TGA GGA TGA 3’, TOXO R2 5’ TCG TCT CGT CTG GAT CGC AT 3’) that duplicates the fragments with length of 200 and 134 bp. The access number of the RE sequence was AF146527 (8). The reaction of PCR was conducted by the thermal cycler system (Bio-Rad Company, USA). The PCR method was performed with an initial denaturation at 95°C for 10 minutes, denaturation at 92°C for 30 seconds, annealing at 55°C for 50 seconds, extension at 72°C for 30 seconds and final extension at 72°C for seven minutes.
2.5. Nested Polymerase Chain Reaction
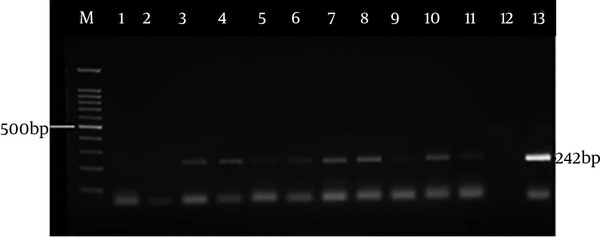
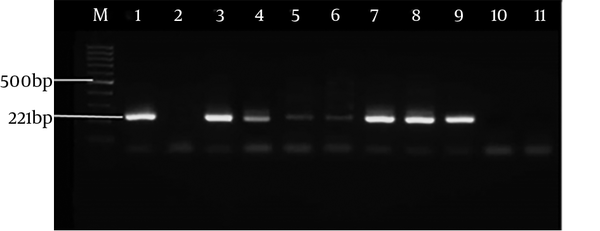
The PCR-positive samples were examined by the nested-PCR method, based on the SAG2 genomic target. In this step, four pairs of primers were used, two of which were utilized as the forward (5’) primers and two pairs as the reverse (3’) primers. The characteristics of the utilized primers for nested PCR are shown in Table 1. Then, the nested-PCR method was conducted in four phases, two phases were for forward (5’) primers and two phases for reverse (3’) primers and the first phase products were used as DNA for the second phase. The length of the multiplied fragment in forward direction (5’) was 242 bp and the length of the multiplied fragment in reverse direction (3’) was 221 bp.
| Sequence | Primer Name | Primer |
|---|---|---|
| 5’ GCT-ACC-TCG-AAC-AGG-AAC-AC 3’ | SAG2.F4 | Forward primer 1 |
| 5’ GCA-TCA-ACA-GTC-TTC-GTT-GC 3’ | SAG2.R4 | Forward primer 2 |
| 5’ GAA-ATG-TTT-CAG-GTT-GCT-GC 3’ | SAG2.F | Forward internal primer 1 |
| 5’ GCA-AGA-GCG-AAC-TTG-AAC-AC 3’ | SAG2.R2 | Forward internal primer 2 |
| 5’ TCT-GTT-CTC-CGA-AGT-GAC-TCC 3’ | SAG2.F3 | Revers primer 1 |
| 5’ TCA-AAG-CGT-GCA-TTA-TCG-C 3’ | SAG2.R3 | Revers primer 2 |
| 5’ ATT-CTC-ATG-CCT-CCG-CTT-C 3’ | SAG2.F2 | Revers internal primer 1 |
| 5’ AAC-GTT-TCA-CGA-AGG-CAC-AC 3’ | SAG2.R | Revers internal primer 2 |
The Primers Utilized in Nested Polymerase Chain Reaction Method
2.6. Restriction Fragment Length Polymorphism and Genotyping
For genotyping of T. gondii at the SAG2 locus, the Sau3A I restriction endonuclease enzyme (Fermentas Company, Germany) in the forward direction and Hha I restriction endonuclease enzyme (Fermentas Company, Germany) in the reverse direction were used. The Sau3A I restriction endonuclease enzyme can cause the separation of type I and type II T. gondii genotypes from the type III genotype.
3. Results
3.1. Demographic Characters of the Patients
Of 170 patients, 74 (43.5%) were female and 96 (56.5%) were male. The mean age of the patients was 6.18 ± 3.22 years and the highest frequency was 28.8%, 28.2%, and 27.1% in the age group of four to six, seven to nine, and one to three years, respectively. In addition, the lowest frequency was 3.5% and 12.4% in the age group of 13 to 15, and 10 to 12 years, respectively. Table 2 shows the frequency of the malignancies in the subjects.
| Malignancy | Frequency (N) | Percentage |
|---|---|---|
| Acute lymphocytic leukemia (ALL) | 116 | 68.2 |
| Lymphoma | 24 | 14.1 |
| Osteosarcoma | 11 | 6.5 |
| Retinoblastoma | 7 | 4.1 |
| Glioblastoma | 10 | 5.9 |
| Colorectal cancer | 1 | 0.6 |
| Peritoneal carcinoma | 1 | 0.6 |
| Total | 170 | 100 |
The Frequency of Malignancies in the Subjects
3.2. Polymerase Chain Reaction Results
Figures 1 and 2 show the electrophoresis of PCR products related to blood and CSF samples, respectively. Of 170 blood samples, seven cases were positive and six and one samples were observed in patients with ALL and lymphoma, respectively. In addition, of 170 CSF samples, 16 cases had positive results; 12, three, and one samples were found in the patients with ALL, lymphoma, and retinoblastoma, respectively. Table 3 shows all positive CSF samples detected in children with immunodeficiency, using the molecular analysis.
| No. | Gender | Age | Type of Cancer | Blood Samples | CSF Samples | Genotype |
|---|---|---|---|---|---|---|
| 1 | Female | 3 | All | + | + | II |
| 2 | Female | 3 | All | + | + | II |
| 3 | Male | 8 | All | + | + | II |
| 4 | Male | 8 | All | + | + | II |
| 5 | Male | 9 | All | + | + | II |
| 6 | Female | 8 | All | + | + | II |
| 7 | Male | 13 | Lymphoma | + | + | Mix of II and III |
| 8 | Female | 2 | All | - | + | II |
| 9 | Female | 4 | Retinoblastoma | - | + | II |
| 10 | Male | 4 | All | - | + | II |
| 11 | Male | 3 | All | - | + | II |
| 12 | Female | 5 | All | - | + | II |
| 13 | Male | 6 | Lymphoma | - | + | II |
| 14 | Male | 8 | All | - | + | II |
| 15 | Female | 11 | Lymphoma | - | + | II |
| 16 | Female | 2 | All | - | + | II |
Positive Cerebrospinal Fluid Samples Detected in Children with Immunodeficiency by the Molecular Analysis (Using RE Gene)
3.3. Nested Polymerase Chain Reaction Results
Figure 3 indicates the electrophoresis of the nested PCR products (242 bp) related to SAG2 locus in forward direction, and Figure 4 shows the electrophoresis of the nested PCR products (221 bp) related to SAG2 locus in reverse direction. Of 16 CSF samples (observed in the PCR method), all cases were found to have positive results by the nested-PCR method while all blood samples were negative.
3.4. Genotyping Results
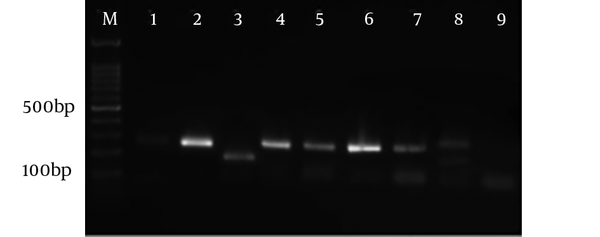
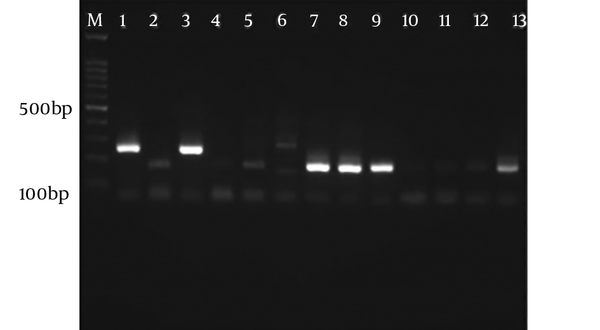
Figure 5 shows the electrophoresis of the RFLP samples using the Sau3A I enzyme in forward direction (5’). Also, Figure 6 demonstrates the electrophoresis of the RFLP samples using Hha I enzyme in reverse direction (3’). After genotyping CSF samples, T. gondii type II was found in 15 samples and one sample was a mix of both types II and III.
Electrophoresis of RFLP samples using Hha I enzyme in reverse direction (3’). M: Marker 100 bp, sample 1: Positive control of genotype I, sample 2: Positive control of genotype II and sample 3: Positive controls of genotype III, the 4 - 13 samples are genotype II according to cutting pattern.
5. Discussion
The results indicated that of 340 specimens, 23 samples (seven blood samples and 16 CSF samples) were detected positive by the PCR method and all CSF samples were approved by the nested-PCR method, using the SAG2 locus while all blood samples were negative. The results demonstrated that the RE gene had a high sensitivity for the recognition of T. gondii DNA in the biological fluids. It has been established that the RE gene has a high sensitivity for the diagnosis of the parasite in biological samples (14). Therefore, the sensitivity of the RE gene is 10 to 100 times more than the B1 gene (15). Accordingly, Fallahi et al. showed that the RE gene could identify the parasite DNA in seronegative samples and the sensitivity of the gene is more than the B1 gene (16). Furthermore, based on the results of the present study, seven blood samples were found positive by the PCR method using the RE gene and these positive samples were observed negative in the nested-PCR analysis, using the SAG2 locus. The failure to amplifying the locus of SAG2 related to T. gondii in blood samples was probably a result of poor sample preservation or too few parasites (12). The current results showed that the sensitivity of the RE gene was more than SAG2 locus for the diagnosis of the parasite in blood samples.
The results showed that after genotyping CSF samples, T. gondii type II was found in 15 samples and one sample was a mix of both types II and III. However, genotype I was not observed. These results were consistent with the studies of How et al. (12) and Mondragon et al. (13), that reported genotype II as the most common genotype. The nested PCR technique at the SAG2 locus is a useful method in evaluating a variety of samples for the genotyping of T. gondii (12). The studies conducted in Iran are very rare. In 2003, Behzadi et al. used the PCR-RFLP technique on human and mouse. They showed that genotype II was the most dominant genotype and genotype I was rarely observed, yet genotype III was not reported (11). Unlike all these studies, Fuentes et al. reported that genotype I was the most common genotype (17). In addition, Zia-Ali et al. investigated different genotypes of T. gondii in domestic birds of Iran and they pointed out that genotype III was the prevailing genotype among birds. In this research, genotypes I and II were not reported (18). Also, Zia-Ali et al. evaluated the different genotypes of the parasite, using the microsatellite technique on different animals, such as sheep, goat, duck, chicken, and cat. They showed that genotype III was the prevailing genotype in Iran yet genotype II was not reported (19).
The strengths of this study was the sample type (CSF obtained from children with immunodeficiency), the good sample size, the use of two molecular techniques, and the genotyping of T. gondii in children with immunodeficiency. In addition, the limitations of the research were the lack of informative clinical data related to the tested patients, the lack of serologic testing as well as failure to follow patients. In conclusion, the findings indicated that the RE target can be considered as a sensitive method for the diagnosis of toxoplasmosis in CSF samples, especially in children with immunodeficiency. In addition, this study showed that the genotype II was the prevailing genotype in patients.