1. Background
Cryptosporidium is a worldwide zoonotic apicomplexan protozoan parasite (1) infecting the gastrointestinal tract of a wide range of vertebrates, including humans, livestock, wild animals, and birds (2, 3). The most common routes for Cryptosporidium transmission are direct fecal contamination or contaminated drink water that result in diarrhea in a variety of vertebrate hosts (4). In humans, this infection is commonly found in children and immunocompromised individuals (5, 6); in children, cryptosporidiosis influences growth, physical activity, and cognitive function. Cryptosporidiosis has also been identified as the leading global cause of diarrheal mortality among infants aged between 12 and 23 months (7, 8).
In normal and healthy individuals, cryptosporidiosis is known as a self-limiting disease (9, 10) lasting, on average, up to two weeks, although the situation differs in immunocompromised individuals. Cryptosporidiosis can be life-threatening since no fully effective drug treatment has been yet developed for it. The primary site of Cryptosporidium infection in humans is the small intestine; however, in immunocompromised individuals, biliary tract, lungs, or pancreas are the most frequently affected extraintestinal organs (8). Recently, molecular studies have disclosed that cryptosporidiosis is caused by at least 15 different species, the most common of which are Cryptosporidium hominis (11, 12) (humans are the only natural host for this species), Cryptosporidium parvum (C. parvum) (which infects bovines as well as humans), Cryptosporidium mulla, Cryptosporidium felis, Cryptosporidium carnivorous, and Cryptosporidium humicus (13).
The prevalence rate of Cryptosporidium varies in different regions of the world; for instance, it is 0.6 to 4.3% in North America, 3 to 10% in Asia, Australia, Africa, and Central America, and 1 to 2% in Europe (5). In Iran, according to the results of a systematic and meta-analysis study, the prevalence rate of cryptosporidiosis among children, healthy people, gastroenteritis patients, and immunocompromised patients has been estimated to be 3.65, 2.94, 1.29, and 4.54%, respectively, using the random effects model. The findings of a phylogenetic analysis inferred by gp60 and 18S ribosomal RNA markers have indicated that highest infection rates belong to C. parvum (particularly, subtype IIaA15G2R1) and C. hominis among understudied groups (14). Considering the fact that there was no comprehensive study on Cryptosporidium in Urmia city, we designed this study to compare the sensitivity and specificity of acid-fast (AF) staining, polymerase chain reaction (PCR), and enzyme-linked immunosorbent assay (ELISA) for diagnosis of Cryptosporidium and its dominant species among children admitted to Motahari Hospital in Urmia city. The intention was to select the most favorable method among the three compared here with low test duration and procedure cost and also with the highest sensitivity in detecting Cryptosporidium, for differential diagnosis of cryptosporidial diarrhea among children and immunocompromised individuals.
2. Objectives
The objective of the study was to evaluate the efficiency of different diagnostic tools for detection of Cryptosporidium in pediatrics with diarrhea based on conventional, ELISA and molecular assays. The aim was also to sequence PCR products to determine epidemiology of Cryptosporidium species (spp.) for the first time in West Azerbaijan, Iran.
3. Methods
3.1. Sample Collection
For this diagnostic medical study, 221 diarrheal stool samples were collected from children admitted to Motahari Hospital, the pediatric-specific center in Urmia city, from October 2017 to March 2018. Demographic features such as age and gender and some other factors including place of residence (city or village), soil contact, animal contact, breast-feeding, stay in kindergarten and family's socioeconomic conditions were applied in the questionnaire. The samples were transferred to the laboratory at Urmia University of Medical Sciences on the same day of collection.
3.2. Evaluation of Stool Samples
Since it was impossible to perform all the three methods simultaneously, we divided each diarrheal stool sample into three parts: one part was kept for detection of Cryptosporidium oocytes by the AF (Ziehl-Neelsen) staining method (221 samples), one part was transmitted into a test tube containing 70% alcohol for the PCR method (221 samples), and one part was maintained in normal saline at -20ºC for the ELISA method (94 samples).
3.2.1. Ziehl-Neelsen Staining Method
All the samples were first examined by the formalin-ethyl acetate sedimentation method (15), as in the following procedure. To each test tube, 7 ml of formalin and 2 - 3 mL of each diarrheal stool were added, followed by adding 3 mL of ethyl acetate. After sealing the tubes, their contents were mixed perfectly using a vortex. Then, the tubes were centrifuged at 450 ×g for 5 minutes. Afterward, from the top to bottom of the tube, four visible layers were formed, including ethyl acetate, fat and stool debris, formalin, and an oocyst containing sediments. After discarding the first three layers, smears were prepared on glass slides for each sample and dried at room temperature. The Ziehl-Neelsen staining method was performed after the slides were prepared and fixed by methanol. In brief, carbol fuchsin was used to stain the slides for 15 minutes, which were then decolorized with 50% alcohol for 3 - 5 seconds. In the next step, the slides were washed with water. Finally, 1% sulfuric acid was used to decolorize the slides until the disappearance of red color. At this moment, the slides were rinsed again with water. After drying the slides, methylene blue was used to stain the field for one minute, which was subsequently washed with water. By observing the slides under immersion oil (objective *100), we identified red Cryptosporidium oocytes with the size range of 4 - 6 microns and black granules on blue background. About 30 minutes was required for the AF staining method to detect Cryptosporidium (15).
3.2.2. PCR Method
The DNA extraction was performed using a commercial kit (Yekta Tajhiz Azema, Iran), following the manufacturer’s instructions for use. For multiplication of the SSU rRNA gene of Cryptosporidium spp., forward (5’GACATATCATTCAAGTTTCTGACC3’) and reverse (5’CTGAAGGAGTAAGGAACAACC3’) primers were designed. The process was carried out by adding 1 μL of each forward and reverse primer, 3 μL of the extracted DNA, 12.5 μL of the master mix, and 7.5 μL of sterile distilled water to the micro-tubes. Amplification of Cryptosporidium DNA was accomplished with a thermocycler device, as described in Table 1.
| Stage | Temperature (°C) | Time (s) | Cycle |
|---|---|---|---|
| Hot start | 94 | 120 | 1 |
| Denaturation | 94 | 120 | 35 |
| Annealing | 58 | 60 | 35 |
| Extension | 68 | 120 | 35 |
| Final extension | 72 | 420 | 1 |
Thermal Cycling Parameters of PCR Primers for Amplification of Cryptosporidium DNA
The PCR product was first loaded on a 1% agarose gel and then electrophoresed using 1% Tris/Borate/EDTA solution for 20 minutes. Finally, digital images were taken using a gel imaging system. Cryptosporidium spp. bands with the length of 830 bp were identified. Positive PCR products were sequenced, and the sequences were visualized using the Chromas software (version 2.6). The sequences were then compared to those registered in the GenBank using basic local alignment search tool (BLAST) software. The time period required for Cryptosporidium DNA extraction and the PCR method was about 8 hours.
3.2.3. ELISA Method
A commercial ELISA kit (Cryptosporidium 2nd Generation [Fecal], USA) was used in the study. To conduct the experiment, a buffer solution was added to all wells of a plate, and then negative and positive controls and (94) samples were added to each well. The plate was incubated at room temperature for 1 hour, and after washing the wells, enzyme conjugate was added to all the wells and incubated at room temperature for 30 minutes. After the washing step, 100 μL of chromogenic solution was added to each well and the plate was incubated at room temperature for 10 minutes. The reaction was stopped by adding 100 μL of stop solution to each well. The optical density of the wells was measured by an ELISA reader (Awareness Stat Fax 2100, Palm city, FL, USA). The optical density above 0.08 was considered as positive for Cryptosporidium antigens (the cut-off point). The time taken to determine the Cryptosporidium antigens by the ELISA method was 2 hours and 30 minutes.
3.3. Ethical Issues
The study was approved by the Ethics Committee of Urmia University of Medical Sciences, Urmia, Iran (approval number: 1396.264.REC.UMSU.IR).
3.4. Statistical Analysis
By considering the PCR method as the gold standard, we compared sensitivity, specificity, and negative and positive predictive values of the AF staining, PCR, and ELISA methods using the McNemar’s chi-squared test (16).
4. Results
The results from the samples analyzed using the three aforementioned methods, as shown in Table 2, revealed that the number of Cryptosporidium-positive samples was four, five, and seven for AF staining, ELISA, and PCR, respectively.
| Method | Number of Samples | Positive Cases, N (%) | Negative Cases, N (%) |
|---|---|---|---|
| Acid-fast staining | 221 | 4 (1.8) | 217 (98.2) |
| ELISA | 94 | 5 (5.32) | 89 (94.68) |
| PCR | 221 | 7 (3.2) | 214 (96.80) |
The Results of the Acid-Fast Staining, ELISA, and PCR Methods in Detecting Cryptosporidium in the Stool Samples
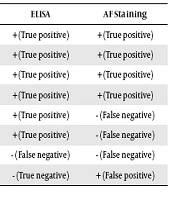
The results also indicated that the ELISA and PCR methods yielded a higher number of Cryptosporidium-positive cases compared to the AF staining method, with the most positive cases being observed using the PCR method. The number of true positive, true negative, false positive and false negative obtained using the ELISA and AF staining methods is shown in Table 3. In this study, the prevalence rate of Cryptosporidium among the studied children was evaluated to be about 3.2%.
| PCR | ELISA | AF Staining |
|---|---|---|
| + | + (True positive) | + (True positive) |
| + | + (True positive) | + (True positive) |
| + | + (True positive) | + (True positive) |
| + | + (True positive) | + (True positive) |
| + | + (True positive) | - (False negative) |
| + | + (True positive) | - (False negative) |
| + | - (False negative) | - (False negative) |
| - | - (True negative) | + (False positive) |
The Number of True Positive, True Negative, False Positive and False Negative Samples in the ELISA and AF Staining Methods
Based on statistical parameters, ELISA and AF (Ziehl-Neelsen) staining were compared with PCR. The parameters of true positive, true negative, false positive, false negative, sensitivity, specificity, positive predictive value, negative predictive value and P value for the AF staining and ELISA methods are presented in Table 4.
| Acid-Fast Staining Method | ELISA Method | P Value | |
|---|---|---|---|
| True positive | 4 | 5 | |
| True negative | 213 | 87 | |
| False positive | 1 | 0 | |
| False negative | 3 | 2 | |
| Sensitivity (%) | 57.14 | 71.4 | 0.724 |
| Specificity (%) | 99.53 | 100 | < 0.001 |
| Positive predictive value (%) | 80 | 100 | 0.579 |
| Negative predictive value (%) | 98.6 | 97 | < 0.001 |
Calculations and Results of the Sensitivity, Specificity, and Positive and Negative Predictive Values of the Acid-Fast Staining and ELISA Methods in Comparison with PCR
A positive correlation was observed with age (< 5 years), place of residence (city or village), and animal contact (P < 0.001). However, there was no statistically significant (P = 0.08) correlation of gender, social-economic condition, breast-feeding, and stay in kindergarten with positivity to parasite.
Based on the Sanger method, five Cryptosporidium-positive isolates obtained by the PCR method were sequenced by Kawsar Biotech Company (Iran). The results identified all the five isolates as C. parvum, which was registered in the GenBank under the accession numbers MK426792, MK426793, MK426794, MK426795, and MK426796.
5. Discussion
In this study, the prevalence rate of Cryptosporidium was obtained as 3.2% among children with diarrhea in Urmia city. This result is relatively in agreement with those obtained from the cities of Hamedan (5.4%), Isfahan (4.6%), Gonbad Kavoos (4.94%), and Tehran (2.40%) (17-20). However, this positive association was not observed between our study and a previous study performed in the cities of Urmia and Nagadeh (7.66%) (21). The probable reason for this difference can be the time, during which the latter study was conducted (27 years ago), as well as levels of awareness, knowledge, and health that have increased among people, thereby resulting in the decreased prevalence rate.
In the current study, three diagnostic methods of Cryptosporidium were compared using stool samples. To select the best standard and cost-effective method in routine diagnosis of this parasite, various factors such as time duration for performing tests, cost, fluency, sensitivity, specificity, and positive and negative predictive values were considered. The results showed that the PCR and ELISA methods were more accurate than the microscopic method. In a similar study, Yilmaz et al. (21) made a comparison between the AF staining and ELISA methods. They detected Cryptosporidium in stool samples of 2000 children in Turkey. According to their results, Cryptosporidium antigens were detected in 97 samples using the ELISA method; however, Cryptosporidium were detected only in 39 samples using the AF staining method. This indicated the higher sensitivity of ELISA relative to AF staining. In another similar study, Morgan et al. (22) compared the AF staining procedure with the PCR test to detect Cryptosporidium among 511 stool specimens referred for screening on the basis of diarrhea. The results showed that PCR and AF staining detected a total of 36 and 29 positive cases among the 511 stool samples, respectively. It was also revealed that the sensitivity and specificity of the AF staining method were 83.7% and 98.9%, respectively, when compared with PCR (100% sensitivity) (23).
In our work, the gold standard was the PCR method. All the negative and positive samples were analyzed by the AF staining method and tested by the PCR method. However, because of some limitations, only 94 samples were selected for the ELISA method. After performing the ELISA test, the sensitivity as well as the positive and negative predictive values of the AF staining method were calculated using PCR, as the standard method. The obtained results showed a lower sensitivity for the AF staining method in comparison with the PCR method. Moreover, the number of false negative samples was observed to be relatively high while using the staining method.
This was the first study that applied molecular methods to determine the prevalence rate of C. parvum in children in West Azerbaijan province, North-West of Iran. In the present study, using PCR, all isolates of the species from children were identified as C. parvum. This result correlates with the Memar et al.’s (23) and Nazemalhosseini et al.’s (24) findings showing that C. parvum is the predominant species found in individuals in Iran. Moreover, similar to our results, Taghipour et al. (19) recognized C. parvum as the dominant isolate among children with diarrhea in Tehran city; based on the sequence analysis of the GP60 gene, 17/19 (89.47%) of the positive isolates were C. parvum and 2/19 (10.52%) were C. hominis (19). Our results contradict with the findings of Jiang et al. (25) who showed C. andersoni and C. hominis as the dominant isolates with regard to the SSU rRNA gene.
Regarding the advantages and disadvantages of the three methods used in this study, the time spent on PCR, ELISA, and AF staining was 8, 2.5, and 0.5 hours, respectively. Further, the cost was extremely high for the PCR method, but the minimum for the ELISA and AF staining methods. Hence, due to the low sensitivity of the staining method and considering its cost, test time, and sensitivity, the ELISA method is more favorable for routine use in the diagnosis of Cryptosporidium among children and immunocompromised people. Since Cryptosporidium-specific antibodies are attached to wells of ELISA plates in this method, it is logical to conclude that the ELISA method is a highly sensitive method and can play an important role in promoting the community health.
5.1. Conclusions
C. parvum was identified as the only infectious agent for humans in the study region. It appears that more investigations are needed to find the infection source in order to design prevention strategies. The most predictive sources for humans are animals and water that should be considered in future studies. Designing a handmade ELISA kit could be the next step to facilitate the routine use for Cryptosporidium detecting. Another aim of this study was to determine Cryptosporidium spp. that affected children; thus, we used PCR as the gold standard to cover all the samples.