1. Background
After more than a century of the discovery of the tuberculosis virus by Robert Koch, the disease remains one of the most common infectious diseases with the highest mortality after the immune deficiency virus (HIV) (1). After 1982 due to medical advances in the discovery of the disease, identification of pathogenesis, and establishment of a standard treatment regimen to combat tuberculosis, there was a belief among physicians that the disease would be limited to case reports and medical literature by 2000. However, the spread of AIDS epidemics, the emergence of Mycobacterium tuberculosis strains with multiple-drug resistance (MDR-TB), and extra-drug resistance (XDR-TB) have caused the disease to be declared in 1993 as a global emergency (2, 3). According to available sources, it was estimated that in 2012, nearly 3.6% of new cases of tuberculosis and about 20.2% of tuberculosis cases were MDR-TB cases, and what was more worryingly was that only less than 25% of MDR-TB cases were identified and reported in that year (4). In addition, according to the World Health Organization (WHO) reports in 2015, around two billion people worldwide are infected with Mycobacterium tuberculosis bacillus, although they do not feel any signs of illness. Moreover, 10.4 million new cases and 1.4 million mortalities due to tuberculosis have been estimated for this year (5). The incidence of tuberculosis in Iran is estimated to be 21 cases per 100,000 people. This rate is much lower than in the neighboring countries such as Afghanistan, Pakistan, Azerbaijan, and Armenia (these countries are among “high TB-burden” countries). Thus, one of the programs for controlling tuberculosis in Iran is to control and monitor immigration to the country. However, the second report regarding tuberculosis resistance to all drugs (XXDR-TB or TDR-TB) after Italy was reported from Iran (1, 3).
Pyrazinamide is considered one of the four first-line anti-tuberculosis drugs within the first two months of treating the disease. The anti-tuberculosis activity of this drug was first discovered in 1952, but its role in accelerating the effect of treatment was found when it was co-administered with isoniazid and rifampin (1, 6). By eliminating the active and inactive forms of tuberculosis bacilli in acidic environments within macrophages, this drug, in addition to shortening the course of treatment, is also very effective in eradicating the organism in the patient’s body (6, 7). The role of pyrazinamide is important in standard tuberculosis treatment regimens because it is more effective in the acidic conditions of macrophages and monocytes against Mycobacterium tuberculosis (1, 6). Today, this drug is the second-line treatment of MDR-TB and XDR-TB patients that eliminates latent tuberculosis bacilli in macrophages and prevents the recurrence of the disease (8). Pyrazinamide is a precursor and is converted to its active form (pyrazinoic acid or POA) by the enzyme pyrazinamidase after entering the cytoplasm of Mycobacterium tuberculosis (Mtb), causing the acidification of the Mtb cytoplasm, inhibiting the synthesis of fatty acids, and disrupting the cell wall function. Pyrazinoic acid can also prevent the translation process by binding to the ribosomal protein S1 (rpsA gene product) (1, 9). As known, PncA gene mutations are one of the major mechanisms of resistance to pyrazinamide, followed by rpsA gene mutations. However, according to recent studies, panD gene mutations may also be involved in this phenomenon. This gene encodes an aspartate decarboxylase enzyme and plays an important role in the metabolism of amino acids alanine and aspartate. Besides, PanD gene mutations are observed in the Mycobacterium tuberculosis strains of the canettii subtype (one of the members of the Mycobacterium tuberculosis complex, which is naturally resistant to pyrazinamide, such as Mycobacterium bovis and Mycobacterium bovis BCG) and MDR-TB strains. It was suggested that the mutations of this gene are related to the resistance of TB bacilli to pyrazinamide, but the required verification is under review and needs more extensive studies (1, 10).
Contrary to anti-tuberculosis drugs, activating pyrazinamide in acidic pH has provided difficulties for the evaluation of this drug. Also, considering that there is currently no standard method for evaluating susceptibility to pyrazinamide, this is usually ignored in mycobacteriology laboratories, especially in developing countries, and most results on determining the anti-tuberculosis drug susceptibility do not include the results regarding pyrazinamide (4, 11). Based on a meta-analysis of the rate of resistance to pyrazinamide, the prevalence of resistance to pyrazinamide in the susceptible isolates to the drug is estimated to be 0% - 9% and 31% - 89%, respectively (12).
Considering previous studies in the city of Isfahan, it was found that the most important factor for TB in this region is due to the reactivation of the previous infection to TB bacillus (Mtb), and it was also revealed that by eliminating the inactive forms of TB bacilli in the lung macrophages, pyrazinamide can prevent the reactivation of tuberculosis (1, 13).
2. Objectives
Due to the rarity of information about resistant strains to Mycobacterium tuberculosis drug as compared to pyrazinamide, this study aimed to estimate and evaluate resistance to pyrazinamide in isolates resistant to Mycobacterium tuberculosis drug in patients in the city of Isfahan.
3. Methods
3.1. Isolation of Mycobacterium tuberculosis Isolates, Culture, and Diagnosis Using Phenotypic and Molecular Tests
In this study, 47 isolates of drug-resistant Mycobacterium tuberculosis bacillus (Mtb) (including MDR, isoniazid mono-resistant, and rifampin mono-resistant strains) were studied which were isolated from patients referring to the Isfahan Tuberculosis Center. After initial culture, these isolates were transferred to the Löwenstein-Jensen medium and underwent phenotypic tests such as the Ziehl-Neelsen method of staining, growth rate, colony morphology, pigment production, biochemical tests including nitrate recovery, niacin production, urease, iron withdrawal, growth on MacConkey agar without crystal violet, hydrolysis of tween 80, three-day Arylsulfatase, quantitative catalase resistant to heat, and cytochrome P450-PCR. Cytochrome P450-PCR was performed according to the primers and temperature programs of Darban-Sarokhalil et al. (14).
3.2. Antibiotic Susceptibility Test
The PZA susceptibility test was performed using the conventional 1% proportional method according to the Canetti et al. method (15). In brief, the Löwenstein-Jensen medium containing 400 mg of pyrazinamide and pH = 6 - 6.5 was prepared. Then, 0.1 mL of 10 - 2 and 10 - 4 concentrations, prepared from an equivalent suspension of McFarland’s turbidity were cultured on the media containing the drug while the Löwenstein-Jensen medium with no drug and pH = 6 - 6.5 was used as the control medium. The culture media was kept at 37°C in the incubator with 5% CO2 for four weeks. Besides, M. tuberculosis H37Rv ATCC 2729 and M. bovis ATCC 19210 were used as the controls of the susceptible strains resistant to pyrazinamide. Moreover, PZAse activity using 7H10 agar Middlebrook medium containing 100 mg of pyrazinamide and 2 g/l of sodium pyruvate was done based on the Wayne method (16).
3.3. DNA and PCR Extraction
Cetyl trimethylammonium bromide (CTAB) was used to extract the Mycobacterium tuberculosis isolates’ genomes as per the Miyata et al. protocol. In general, after deactivating the bacteria at 80°C in 20 min, the top solution was separated with the centrifugal force (4,500 rpm for 10 min.) and increased to 10 mL by the TE IX buffer. By re-centrifugal action and adding 100 µL of the lysozyme enzyme and 450 µL of TE IX buffer to the deposit, it was placed at 37°C for 24 h. Next, 72.5 µL of SDS 10% solution and K proteinase solution were added to each sample at 65°C for one day. Each sample was added 100 µL of NaCl at a concentration of 5 mol and 100 µL of CTAB-NACL and placed after vortex at 65°C for 10 min. To each microtube, 750 µL of chloroform isoamyl alcohol mixture was added and after centrifugation, 600 µL of isopropanol was added per 1000 µL of the solution to the deposit of each sample and kept for 24 h at -20°C. Then, 1000 µL of alcohol 70% was added to the deposit, and after centrifuging, drying, and adding 30 µL of TE IX buffer at 37°C for 3 - 4 h, the DNA density was measured by a spectrophotometer.
Then, for the reproduction of the 720-bp fragment containing the complete sequence of the pncA gene, primers P1 (5’-GTCGGTCATGTTCGCGATCG-3’) and P6 (5’-GCTTTGCGGCGAGCGCTCCA-3’) were used as suggested by Scorpio et al. (17) Also, to study mutations in the region “742” pair of the genes from the early part of the rpsA gene, rpsA-1 primers (5’-ATCTGTCCCTACGACCCAAC-3’) and rpsA-5 (5’-ACCGTCCACACCGCCGAGA-3) were used according to Akhmetova et al. (18). The temperature program in Table 1 was used for the reproduction of the genes. Finally, PCR products were sequenced using the aBi3730Xl Automatic Sequencer from Macrogen Co. (South Korea).
3.4. Statistical Analysis
We used chi-square and Fisher Exact tests (P-value < 0.05) in SPSS ver. 20 for comparing the relative frequency of the mutations in the pncA and rpsA genes of Mtb resistant isolates and its types among three resistant groups to isoniazid, rifampin, and MDR (19).
3.5. Nucleotide Sequence Accession Numbers
The sequences containing new mutations, obtained in the present study, were recorded in the GenBank database under access codes MG956806, MG920781, and MG920781 for the pncA gene and access codes MG888046 and MG77745 for the rpsA gene. Additionally, the accession numbers of the sequences of pncA, rpsA, CYP 141, hsp65, and 16S rRNA genes for suspected isolates to the new subtype of Mycobacterium tuberculosis complex are MG920781, MG888046, MH020184, MG994961, and MH002343, respectively.
4. Results
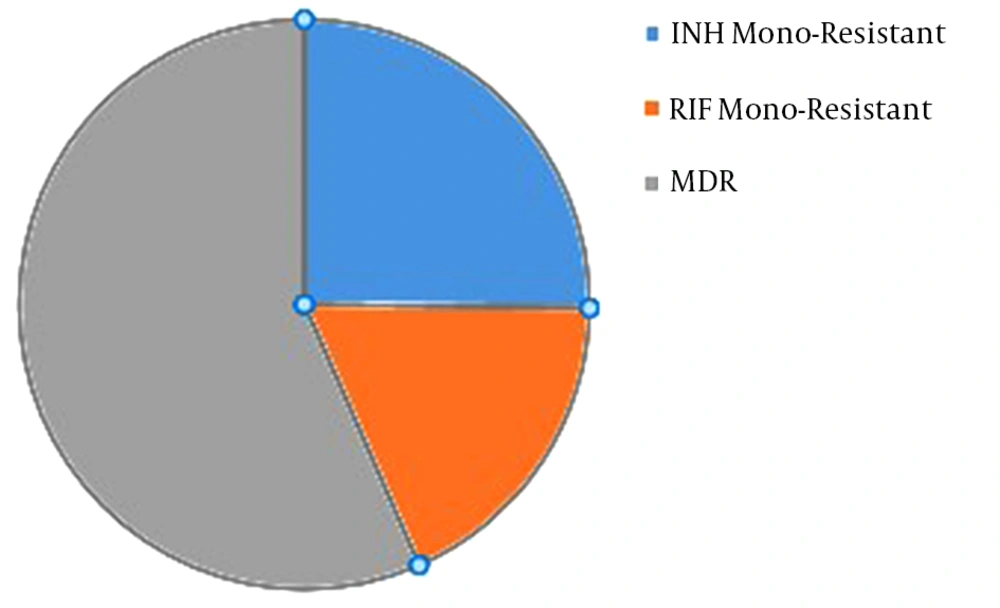
In this study, 47 cultures resistant to Mycobacterium tuberculosis were studied. They included nine (19.1%) MDR isolates, 26 (55.3%) isolates resistant to isoniazid, and 12 (25.5%) isolates resistant to rifampin. Of the patients, 31 (65.9%) cases were men, and 16 (34%) cases were women; 33 (70.21%) cases were Iranian, while 14 (29.7%) cases were Afghani. All isolates were identified as Mycobacterium tuberculosis using the results of phenotypic and molecular tests. Also, based on the results of the proportional and pyrazinamidase test, 19 isolates were resistant to pyrazinamide (Table 2).
| Gene | Primary Denaturation | Repetition | Final Expansion | Cooling | ||
|---|---|---|---|---|---|---|
| Denaturation | Annealing | Expansion | ||||
| pncA | ||||||
| Temperature | 94 | 94 | 55 | 72 | 72 | 12 |
| Time | 15 min | 30 sec | 30 sec | 1 min | 7 min | 10 |
| Course | 1 | 30 | 1 | 1 | ||
| rpsA | ||||||
| Temperature | 96 | 95 | 62 | 72 | 72 | 12 |
| Time | 3 min | 10 sec | 10 sec | 40 sec | 3 min | 10 |
| Course | 1 | 38 | 1 | 1 | ||
| Sample Test | MDR | Rifampin | Isoniazid |
|---|---|---|---|
| Pyrazinamidase | 4 | 10 | 21 |
| Proportional method | 7 | 3 | 9 |
Accuracy of DNA amplification was confirmed by electrophoresis on a 1% agarose gel (Figure 1). Then, PCR products were purified and sequenced using a PCR product purification kit (Roche). The sequences were finally sequenced using MEGA 7.0 software. Sequencing of the 720-bp fragment from P1 primers (from position -150 bp upstream of the starting codon of pncA) and P6 (from the position of + 60 bp downstream of the codon of pncA endpoint) was performed using a pyrazinamide-sensitive Mycobacterium tuberculosis strain (Rv2043c). The analysis of the pncA sequence showed that 16 (84.2%) isolates were eligible to mutate in their pncA sequence (Table 3) while three isolates lacked any mutation. Furthermore, based on the results of the sequence analysis of the primordial 742 bp segment of the rpsA gene and the control strain sensitive to pyrazinamide (Rv2043c), six (14.1%) isolates had mutations in the region of the rpsA gene (Table 3). Three isolates had two types of mutations in the pncA gene. An isolate was found in both the pncA and rpsA genes. An isolate had two “missense” mutations in codons 24 and 524 of the pncA gene, and an isolate had a “silent” Missense mutation and a “missense” mutation in codons 195 and 535 of the pncA gene, respectively. We also investigated three isolates susceptible to all drugs and one isolate of M. tuberculosis H37Rv ATCC 27294 as controls that lacked any mutations in the pncA gene and the primary part of their rpsA.
| Number of Isolates | Resistance to Pyrazinamide | Changes in Nucleotide | Amino Acid Changes |
|---|---|---|---|
| Changes in pncA Nucleotide | |||
| 3 | R | C(195) → t | Ser(65) → ser |
| 1 | R | C(24) → A | Asp(8) → Glu |
| 1 | R | T(184) → C | Pro(62) → ser |
| 1 | R | A(535) → C | Ser(179) → Arg |
| 2 | R | A(535) → G | Ser(179) → Gly |
| 1 | R | C insertion in nt 559 | Frameshift |
| 1 | R | T → G | Stop → Gly |
| 1 | R | GC insertion in nt 444,45 | Frameshift |
| 1 | R | A(139) → G | Thr (47) → Ala |
| 1 | R | T(524) → C | Met(175) → Thr |
| 1 | R | C(185) → A | Pro(62) → His |
| 1 | R | A(410) → C | His(137) → Pro |
| 1 | R | C insertion in 407,8 | Frameshift |
| 1 | R | T(56) → C | T76 → proline |
| 1 | R | T(254) → C | Lu85 → Pro |
| Changes in rpsA Nucleotide | |||
| 1 | R | T insertion 37 | Phenylalanine |
| 1 | R | C insertion 54 | Leu |
| 1 | R | G(618) → A | Leu(206) → leu |
| 1 | R | G(619) → A | Ther(207) → Leu |
| 1 | R | G(221) → A | His(14) → Arg |
| 1 | R | C insert in 40 | Ser |
Of the mutations mentioned, five mutations were new in the pncA and rpsA genes, and are first reported in this study. In addition, one of the isolates under study with high resistance to pyrazinamide (MIC 1200 µg/mL) and significant genetic differences from M. tuberculosis H37Rv ATCC 27294 was considered as a candidate for the new subtype of Mycobacterium tuberculosis native to this geographical area, which requires more research.
Based on the statistical analysis, it was found that the level of pyrazinamide-resistant isolates increased significantly with the increased resistance to isoniazid and MDR-TB (P ≤ 0.001). Generally, the resistance to pyrazinamide was evaluated among the isolates of MDR (77.7%; seven isolates), isoniazid (34.6%; nine isolates), and rifampin (25%; three isolates) (Figure 1).
5. Discussion
One of the most important measures for the proper treatment of tuberculosis patients is the determination of the patient’s drug sensitivity profile before the onset of treatment, which today seems to be necessary in societies due to the increase in cases of drug-resistant tuberculosis (19). Pyrazinamide is one of the first-line drugs for the treatment of tuberculosis that can reduce the duration of treatment from 12 months to six months by eliminating the latent bacilli in the alveolar macrophages. The importance of this drug was further enhanced by the latest WHO recommendations for the use of this drug for the treatment of MDR-TB (6, 8). However, susceptibility to this drug is one of the main challenges of mycobacterial laboratories. This drug requires acidic pH for activity, while the optimum growth rate for Mycobacterium tuberculosis occurs at a neutral pH limit. The BACTEC Mycobacteria growth indicator tube (MGIT) 960 system and 100 µg/mL pyrazinamide concentration are selected nowadays as a reference method for measuring susceptibility to pyrazinamide, but, the results of the false resistance from the inoculation of excess bacteria and the presence of bovine serum albumin is a serious threat to this technique. Moreover, this technique is costly and cannot be performed in many developing countries (6, 20). Molecular methods are considered one of the fastest and least costly methods for measuring the drug susceptibility to Mycobacterium tuberculosis. Based on evaluations and comparisons of the results of phenotypic and molecular methods, it can be safely said that sequencing is one of the most reliable methods for screening pyrazinamide resistance strains (17).
This study was done regarding the limited research and information on the resistance of DR-TB isolates (TDR-TB, XDR-TB, MDR-TB, RIF mono-resistant, and INH mono-resistant) to pyrazinamide. This is the first report of resistance to pyrazinamide among DR-TB isolates from Iran. In this study, pyrazinamide-resistant DR-TB isolates were identified using the proportion method and pyrazinamidase test. Then, mutations of pncA and rpsA genes in the isolates resistant to pyrazinamide were studied using the sequencing technique. In general, 19 isolates from the study were resistant to pyrazinamide, with 16 isolates with mutations in the pncA gene, six isolates having mutations in the rpsA gene, and three isolates lacking any mutation in their sequence. Finally, the resistance to pyrazinamide was evaluated among the isolates of MDR as 77.7% (seven isolates), isoniazid 34.6% (nine isolates), and rifampin 25% (three isolates).
In recent years, the emergence of multi-drug resistant TB has been the greatest threat for TB control programs (21, 22). According to the WHO tuberculosis report, the overall prevalence of Iranian MDR-TB cases is from 1% - 2% (in new cases) to more than 29% (in treatment failure cases) (23). In a recent eight-year prospective study on Iranian TB cases, the average prevalence of MDR-TB cases was reported as 14.18% that was significantly higher than the global average (3.5%) (24). Haratiasl et al. (25) in 2020 showed that the prevalence of pyrazinamide resistance is high among Iranian MDR-TB patients. Several studies have suggested that the frequency of the Beijing family in MDR-TB strains is significantly more than that of other lineages (26, 27). Vaziri et al. (28) in 2019 revealed that MDR-TB strains have several mutations that cause primary resistance to various anti-tuberculosis drugs. We also found that the frequency of resistance to pyrazinamide was significantly higher than that of mono-resistance in TB strains that confirm previous Iranian reports (25, 29).
The results of this study are similar to other studies in this area. For example, in a study by Whitfield et al. (6) on South African pyrazinamide-resistant DR-TB isolates, it was found that pyrazinamide resistance was high among the isolates of MDR-TB (39.3%) and RIF mono-resistant (7.5%). In contrast to our results, the resistance to pyrazinamide is relatively high among rifampin-resistant TB isolates compared than MDR-TB as well as isoniazid-resistant TB isolates., which can be attributed to genetic differences in pyrazinamide-resistant Mycobacterium tuberculosis and transmission of these strains among patients in different countries (30). In a study in India, resistance to pyrazinamide in MDR-TB, INH mono-resistant isolates, and MIO-resistant RIF was 63.7%, 97.2%, and 87.7%, respectively. In the studies from China, Japan, Thailand, and South Africa, pyrazinamide resistance among MDR-TB isolates was in the range of 50% - 60% (19). Briefly speaking, by reviewing different studies, it can be concluded that the prevalence of pyrazinamide resistance in DR-TB isolates, especially MDR-TB, is high. Therefore, pyrazinamide susceptibility testing is essential for these patients before the administration of the drug.
In this study, there were differences between the results of the proportion method and pyrazinamide administration; 19 isolates were identified by the method of proportion as the isolates resistant to pyrazinamide, of which seven isolates were with pyrazinamidase activity. According to similar studies, it can be stated that the test results of pyrazinamidase assay are not reliable, and this test is not an accurate indicator for the screening of pyrazinamide-resistant isolates. According to Singh et al. (31), it was found that the specificity of the BacT/ALERT 3D System and pyrazinamidase assay methods was 100% and 82.85%, respectively, compared to the results in “proportion method LJ”. Another study also found that some Mycobacterium tuberculosis isolates that did not have pyrazinamidase activity did not have any mutations in their pncA gene sequence (32). In the present study, the sensitivity rates of pyrazinamidase and sequencing methods were 63.1% and 84.2%, respectively, in comparison with the proportion method, and the specificity rates of these two methods were 80% and 90%, respectively.
The pncA gene contains 561 nucleotides, which is an encoder of pyrazinamidase enzyme (PZAase) that consists of 186 amino acids (33). Based on various studies, about 90% of the Mycobacterium tuberculosis isolates resistant to pyrazinamide (33)have mutations in their pncA gene, and 5% have mutations in the ribosomal binding site. Mycobacterium bovis and M. bovis BCG are resistant to pyrazinamide due to the alteration of their nucleotide 169 (Asp57 → His) of the pncA gene, and this property has caused the ability to differentiate Mycobacterium tuberculosis from them (34, 35). Mutations of pncA genes are nowadays considered as the main factor in resistivity to pyrazinamide, and sequencing of this gene is one of the fastest and most reliable methods of determining susceptibility to pyrazinamide (36). According to Sreevatsan et al. (35), nucleotide replacement is the most common type of mutation in the isolates of Mycobacterium tuberculosis resistant to pyrazinamide. Also, in this study, as reported by Scorpio and Zhang, mutations in the pncA gene of the isolates resistant to pyrazinamide are spread throughout the gene, in contrast to the rpoB gene that majority of (about 96%) rifampin-resistant Mtb isolates has mutation in the region 27 of the amino acid in the β subunit of the RNA enzyme. (35). The results of our study are consistent with that statement. Moreover, Proline is thought to be one of the most important amino acids in the functional domain of the pyrazinamidase enzyme, with variations in the amino acid content that alter the shape and function of the enzyme and as a result, causes resistance to pyrazinamide (37). In our study, several isolates also have changes in proline replacement. According to previous studies, synonymous mutations are supposed to occur due to the transformation of the predecessors of the Mycobacterium tuberculosis complex (38). In the present study, three isolates had synonymous mutations in nucleotide 195 (C (195) → T). It has also been suggested that one of the most important characteristics of the W family (one of the most important linages of the Mycobacterium tuberculosis complex distributed throughout the world which is resistant to many first-line drugs and some second-line drugs) is “missense” mutation (Thr(47) → Ala) in the pncA gene as one of the most important characteristics of this family. In this study, one of the isolates had the considered mutation (35).
The rpsA gene encodes a ribosomal S1 protein in the 30S component, and the sequence of this gene is protected so that it is considered an option for the differentiation and identification of mycobacteria (1, 39, 40). This gene was first introduced by Shi et al. (9) as a secondary candidate for resistance to pyrazinamide. On the one side, the mutations of this gene have been seen in a limited number of Mycobacterium tuberculosis isolates resistant to pyrazinamide. On the other side, in the study by Alexander et al. (41), none of the pyrazinamide-resistant isolates in the rpsA gene sequence had mutations, but instead, a mutation (A364G) was observed in 13 isolates susceptible to pyrazinamide. This contradictory data cause ambiguity in this regard (41). However, six mutation types were observed in pyrazinamide-resistant isolates in the present study, while drug-sensitive isolates and pyrazinamide-susceptible strains did not have any mutation. Also, the sequence of pncA and rpsA genes of three studied isolates, which were evaluated based on the proportion method for pyrazinamide resistance, lacked any mutations, indicating that the mechanisms of resistance to pyrazinamide are diverse.
5.1. Conclusions
Based on the results of this study, it was found that resistance to pyrazinamide in MDR-TB isolates is high in patients in Isfahan and the evaluation of sensitivity to pyrazinamide is essential for the patients affected by MDR-TB. The present study was the first to measure resistance to pyrazinamide among isolates resistant to Mycobacterium tuberculosis in Iran. Determining the prevalence of pyrazinamide resistance in this category requires a much higher comparison of phenotypic and genotypic methods.