1. Background
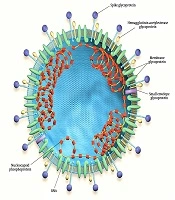
Coronaviruses (CoVs) are positive-sense single-stranded RNA viruses with the largest known genomes among RNA viruses (27 - 34 kb). CoVs constitute the subfamily Orthocoronavirinae, in the family Coronaviridae. The name coronavirus is derived from the Latin word corona, meaning crown or halo, which can be seen under an electron microscope (Figure 1) (1).
2. Coronavirus History
Several studies have been conducted to identify the causes of respiratory tract infections. The severe acute respiratory syndrome coronavirus (SARS-CoV) and human coronavirus NL63 (HCoV-NL63) have proved as two main novel causes of respiratory tract infections (2). CoVs cause acute respiratory infection in humans and also account for about 5% - 30% of total respiratory infections. SARS-CoV caused an epidemic in 2002 - 2003 leading to the severe acute respiratory syndrome in 8000 subjects and 750 deaths. The HCoV-NL63 is classified in group 1 coronaviruses, while SARS-CoV is in group 2 (3).
Acute respiratory infection by SARS-CoV was reported previously due to the mutation or recombination of human and animal CoV. On February 28, 2003, Doctor Urabani from the Private Vietnam French Hospital reported a public threat and requested an immediate emergency meeting to detect this unknown threat. Throughout the investigation, he was infected and died on March 18, 2003. After 18 days, specialists from World Health Organization (WHO) and centers for disease control and prevention (CDC) identified the SARS-CoV as a causative agent for that outbreak (4).
On December 2019, several cases were infected with a novel coronavirus (coronavirus infection disease-19 [COVID-19]) leading to pneumonia in Wuhan, China. Four cases with pneumonia of unknown etiology were reported on December 29, 2019, that herald a similar outbreak with characteristics similar to SARS-CoV in 2003 (5).
3. COVID-19 in Iraq
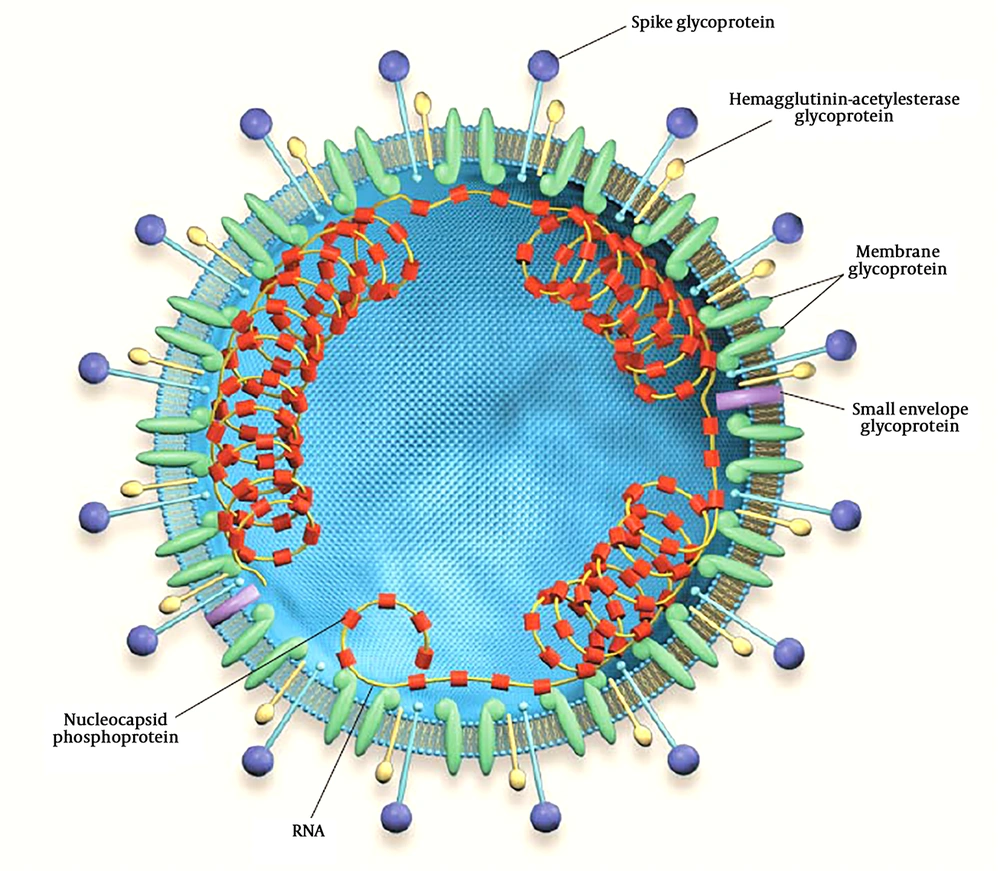
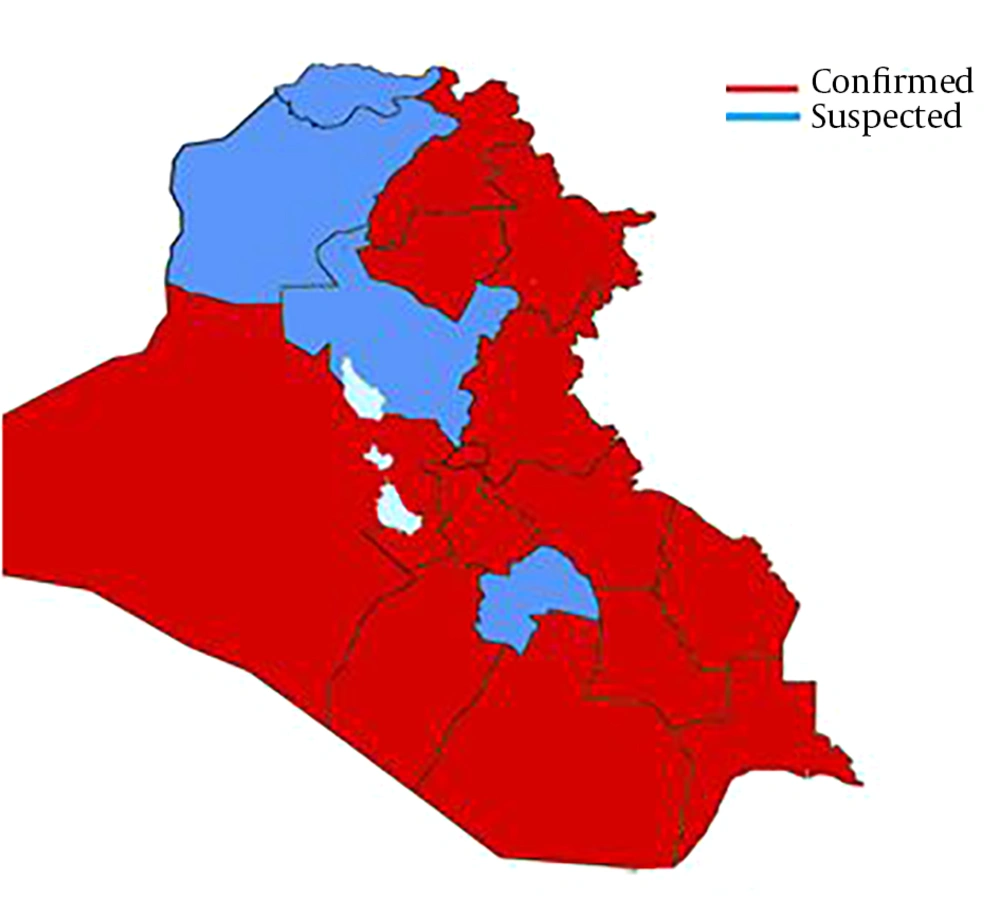
Due to the business relationships between Iraq and China and other infected countries, like Iran, a suspected case of 2019-nCoV was declared on February 22, 2020, in Dhi Qar Province, which later confirmed by a local medical center. Although this finding was denied by the Official Ministry of Health, on February 24, 2020, the first case infected with 2019-nCoV in Najaf was reported, who was a religious student coming from Iran (6). Furthermore, on February 29, 2020, 8 cases with 2019-nCoV were confirmed. On March 4, the Iraqi Ministry of Health declared the first death in Kurdistan and second in Baghdad. The mortality rate was increased; on March 9 the seventh death was declared and the 2019-nCoV-related deaths increased to 67 cases (7). The distribution of suspected and confirmed cases of COVID-19 in Iraq is illustrated in Figure 2.
To date, the confirmed cases are 74, with 7 deaths and 3 recovered cases; therefore, the death rate is 9.45%. On March 11, the number of deaths reached to 8 cases with 79 confirmed cases. Thus, COVID-19 has rapidly increased more than expected (Figure 3) reflecting poor health control.
4. Potential Anti-COVID-19 Treatment
No vaccine or effective antiviral agents have approved for treatment or prevention of the infection with 2019-nCoV. Therefore, the emergence of 2019-nCoV is regarded as a priority by the WHO, CDC, and health agencies for the development of an effective therapeutic agent against this virus. In general, 2019-nCoV as other positive-sense RNA viruses is characterized by high genetic plasticity and mutations due to short replication time, and a higher rate of recombination. These characteristics are challenging and limiting for the discovery of novel antiviral agents (7). However, nucleotide/nucleoside analogue inhibitors (NIs) may be effective agents by reducing the genetic mapping of 2019-nCoV (8). NIs, such as ribavirin, remdesivir, and beta-D-N4-hydroxycytidine can be regarded as promising agents despite the emergence of rapid resistance (9).