1. Background
Human rotavirus infection is associated with severe diarrhea in children and accounts for 39% of hospitalizations. Children below the age of two years are most commonly affected (1).
The viral VP4 and VP7 proteins are the determinants of the serotype specificity of group A rotaviruses. VP4 protein is a non-glycosylated protease-sensitive protein and is thus designated P, whereas VP7 protein is a glycoprotein that determines the G-type of the virus (2). Human genotypes G1P[8], G2P[4], G3P[8], G4P[8], G9P[6], and G9P[8] are responsible for most rotavirus infections among children less than five years of age worldwide (3, 4).
There are two live attenuated oral rotavirus vaccines, the monovalent Rotarix vaccine (GlaxoSmithKline Biologicals, Belgium) with a monovalent G1P[8] strain (5), and the pentavalent human-bovine reassortant RotaTeq vaccine (Merck Co., Inc., United States) that contains rotavirus types of VP7 of G1, G2, G3 and G4, and VP4 of P[8] (6). Both vaccines have been proven to decrease hospitalizations due to severe rotavirus-associated gastroenteritis (7, 8). However, the protection provided by either vaccine may be compromised by infection with rotavirus of G and P types that are not included in the vaccine (9, 10). Moreover, VP4 and VP7 proteins of rotavirus strains that belong to the same G and P serotypes, may display antigenic variations, which define new viral subgenomic lineages or variants that may escape immunity induced by the vaccine (4, 11-13). Evidence was also reported on intergenotypic recombination in VP4- and VP7-coding genes that may play a role in immune evasion (14).
To understand a potential shifting trend following vaccine introduction, rotavirus types circulating before the implementation of rotavirus vaccination program should be identified. A study conducted in India on more than 7,000 rotavirus-positive stool samples collected before the introduction of the oral rotavirus vaccines, ROTAVAC and ROTASIIL, reported the circulation of more than 15 different genotypes, including G1P[8] (38.7%), G2P[4] (12.3%), G9P[4] (5.8%), G12P[6] (4.2%), G9P[8] (4%), G12P[8] (2.4%), and G3P[3] (1.9%) (15).
The last epidemiological study in Kuwait on rotavirus infection dated back to 2005 - 2006, showing the predominance of rotavirus VP7 G1 type (63.8%) in 69 children with severe diarrhea admitted to three major tertiary referral hospitals in Kuwait (16). Rotavirus genotypes G2, G3, G4, and G9 could be detected in 7.3, 4.4, 7.3, and 10.2% of children, respectively. VP4 P[8] was the most prevalent type (89%), followed by P[4] (9%), and P[6] (2%). G1P[8] was the most frequent combination detected in around 63% of patients (16). At that period, the rotavirus vaccine was not available in Kuwait.
2. Objectives
At the end of 2017, the RotaTeq vaccine was included in the Kuwait national immunization program, in a three-dose schedule at 2, 4, and 6 months of age. This led us to investigate, in a retrospective study, the prevalence of rotavirus genotypes circulating in a major tertiary referral hospital in Kuwait during the year 2016, just before the introduction of the RotaTeq vaccine, and to compare the VP4/VP7 subgenomic lineages of circulating rotavirus strains to those of Rotarix and RotaTeq vaccines.
3. Methods
3.1. Ethics Statement
Written informed consent was obtained from the legal guardian prior to the study. The study followed the ethical standards recommended by the Ethical Decision Committee of the Research Administration, Faculty of Medicine, Kuwait University.
3.2. Study Population
Clinical specimens from the Mubarak Al-Kabeer hospital, a major tertiary referral hospital in Kuwait, were sent to the Virology Unit of the Department of Microbiology, Faculty of Medicine, Kuwait University, for routine daily screening for viruses. From each patient, a single fresh stool sample was collected in a clean, dry screw-top container and kept at -80°C till further use. The study population consisted of 101 infants and children under five years of age (47 males and 54 females) with severe acute gastroenteritis. Severe acute gastroenteritis was defined according to the modified Vesikari score (MVS ≥ 11) (17). Severe diarrhea is defined as more than four bowel movements per day for more than three days. The study period was one year, between 1 January and 31 December 2016.
3.2. Rotavirus Detection and Typing
Total RNA was extracted from 200 µL of 10% stool suspension (50 mg of collected feces resuspended in 500 µL phosphate-buffered saline, PBS) using an automated MagNa Pure LC 2.0 system (Roche Diagnostic Systems, Branchburg, NJ, USA). Two reverse transcription-polymerase chain reactions (RT-PCR) were carried out, one to amplify a 905 bp of VP7-coding gene (G-typing) using 9con1 and 9con2 primers (18, 19), and one to amplify an 876-bp VP4-coding gene (P-typing) using con2 and con3 primers (20). PCR products were purified using a Wizard SV GEL and PCR Clean-Up System kit (Promega Corporation, Madison, USA), and the nucleotide sequences of both DNA strands were then determined by performing direct double-strand DNA cycle sequencing using the ABI PRISM® BigDye® Terminator Cycle Sequencing v3.1 kit (Applied Biosystems, Foster City, CA, USA), on an ABI 3500 Genetic Analyzer (Applied Biosystems). Phylogenetic trees were constructed using the neighbor-joining method, with evolutionary distances computed using the maximum composite likelihood method. A bootstrap test with 1,000 replicates was used to estimate the confidence of branching patterns in the trees. The accession number of each rotavirus reference sequence was added to the taxon label in the phylogenetic tree. VP4 and VP7 subgenomic lineages were identified as described previously (13). Rotavirus sequences resulted from this study were uploaded to the NCBI GenBank database (accession numbers MF346890 to MF346925).
3.3. Statistical Analysis
The results were expressed as percentage, median or interquartile range (IQR). A 2x2 or 2xk contingency table was generated to compare different proportions using the chi-square test or Fischer's exact test. Age difference between different groups was assessed using the Kruskal Wallis test. Statistical analysis was performed using IBM SPSS Statistics for Windows, version 25 (IBM Corp., Armonk, N.Y., USA).
4. Results
Rotavirus dsRNA was detected in fecal samples from 25 (24.7%) children between February and May, with a peak in March. All rotavirus cases were admitted to pediatric casualty wards with a diagnosis of severe diarrhea. The median age was one year (range: 1 to 36 months; IQR: 7 to 18 months). Most cases (n = 19, 76%) were children under 2 years of age. Sixty percent of rotavirus cases were males, but there was no significant difference in the distribution of rotavirus positivity by sex (P-value = 0.185).
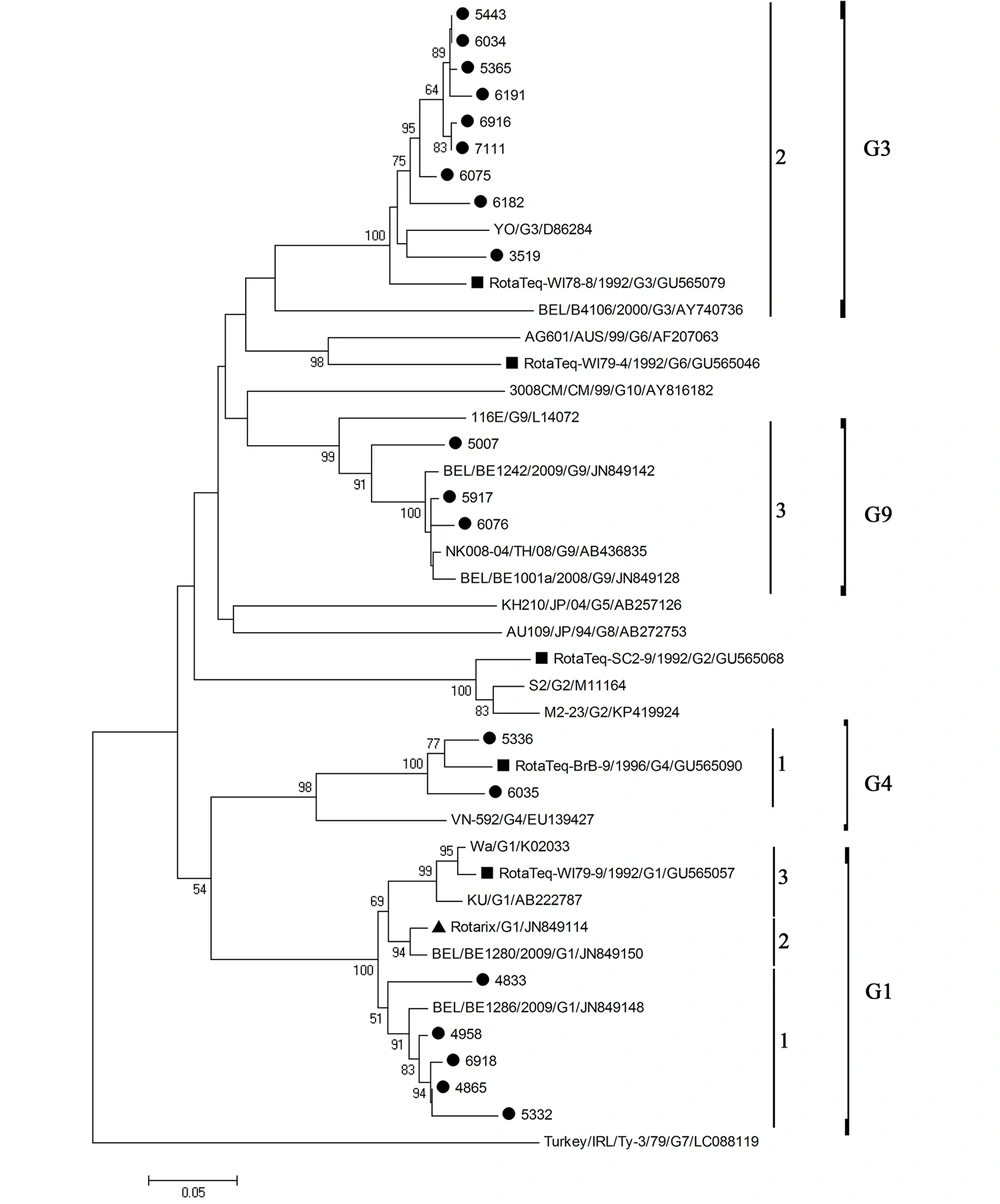
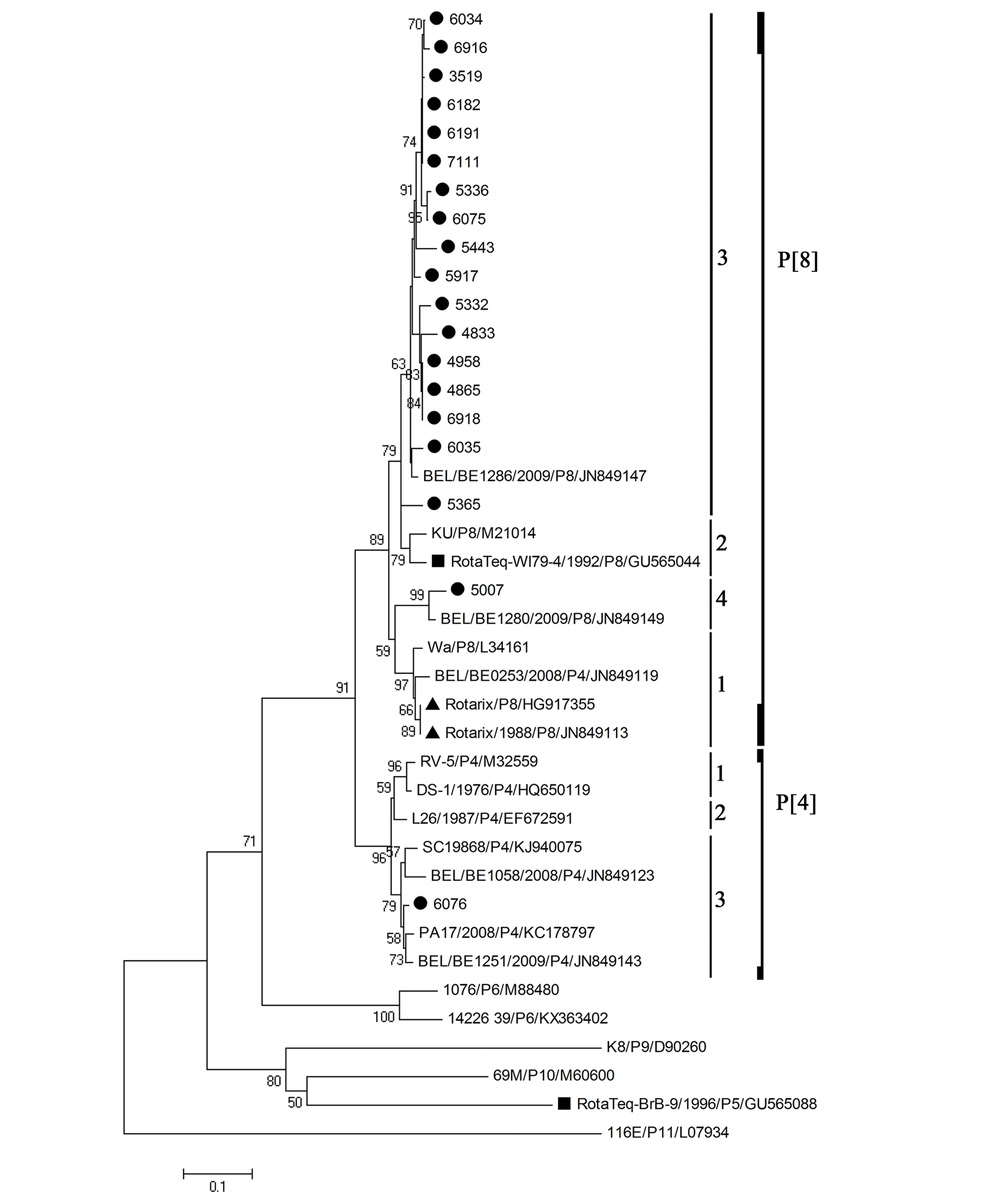
Nucleotide sequence information was obtained for 19 out of 25 rotavirus cases. According to the VP4 and VP7 phylogenetic analyses (Figures 1 and 2), the most prevalent rotavirus type was G3P[8] (n = 9, 47%), followed by G1P[8] (n = 5, 26%), G9P[8] (n = 2, 10.5%), G4P[8] (n = 2, 10.5%), and G9P[4] (n = 1, 5%). Some strains formed clusters with strong bootstrap support (> 90%). However, no identical rotavirus VP7 or VP4 sequences could be identified. The nucleotide sequence identities in the partially amplified VP7 region between different strains were 91 - 98% for G1 type, 89 - 99% for G3 type, 95% for G4 type, and 92% for G9 type, whereas the nucleotide sequence identities between different strains of P[8] type in the partially amplified VP4 region were between 85 and 99%. There was no significant difference in the distribution of rotavirus G types between males and females (P-value = 0.69). In addition, the median age of the children did not vary among different rotavirus G types (P-value = 0.99).
Phylogenetic analysis based on the partial VP7 sequence (426 bp) of human rotavirus A. The black circles next to the taxa represent rotavirus sequences from this study, the black triangle represents the VP7 sequence of the Rotarix monovalent vaccine, while the black squares represent the VP7 sequences of the RotaTeq reassortant vaccine.
Phylogenetic analysis based on the partial VP4 sequence (331 bp) of human rotavirus A. The black circles next to the taxa represent rotavirus sequences from this study, the two black triangles represent the VP4 sequences of two different lots of the Rotarix monovalent vaccine, while the black squares represent the VP4 sequences of the RotaTeq reassortant vaccine.
None of the VP7 and VP4 nucleotide sequences of rotavirus strains were identical to those in Rotarix and RotaTeq vaccines. All VP7 nucleotide sequences of rotavirus G1 type clustered in lineage 1 compared to lineage 2 for Rotarix VP7, and lineage 3 for RotaTeq VP7, with nucleotide sequence identities to Rotarix and RotaTeq VP7 ranging from 91.1 to 95.8, and 88.3 to 91.3%, respectively (Figure 1). VP7 nucleotide sequences of rotavirus G3 type clustered in the same lineage 2 of RotaTeq VP7, with nucleotide sequence identities to RotaTeq VP7 ranging from 88.5 to 93.4%. VP7 nucleotide sequences of rotavirus G4 type clustered in the same lineage 1 of RotaTeq VP7, with nucleotide sequence identities to RotaTeq VP7 ranging from 92.7 to 95.4%. VP7 nucleotide sequences of rotavirus G9 strains clustered in lineage 3. All but one VP4 nucleotide sequences of rotavirus P[8] type clustered in lineage 3, compared to lineage 1 for Rotarix VP4, and lineage 2 for RotaTeq VP4. The nucleotide sequence identities to Rotarix and RotaTeq VP4 ranged from 89.6 to 91.9%, and 89.9 to 94.9%, respectively (Figure 2). The VP4 nucleotide sequence of the strain 6076 clustered in P[4] lineage 3 and showed 83.5% nucleotide sequence similarity to Rotarix VP4 (P[8]) and 82.6% to RotaTeq VP4 (P[8]).
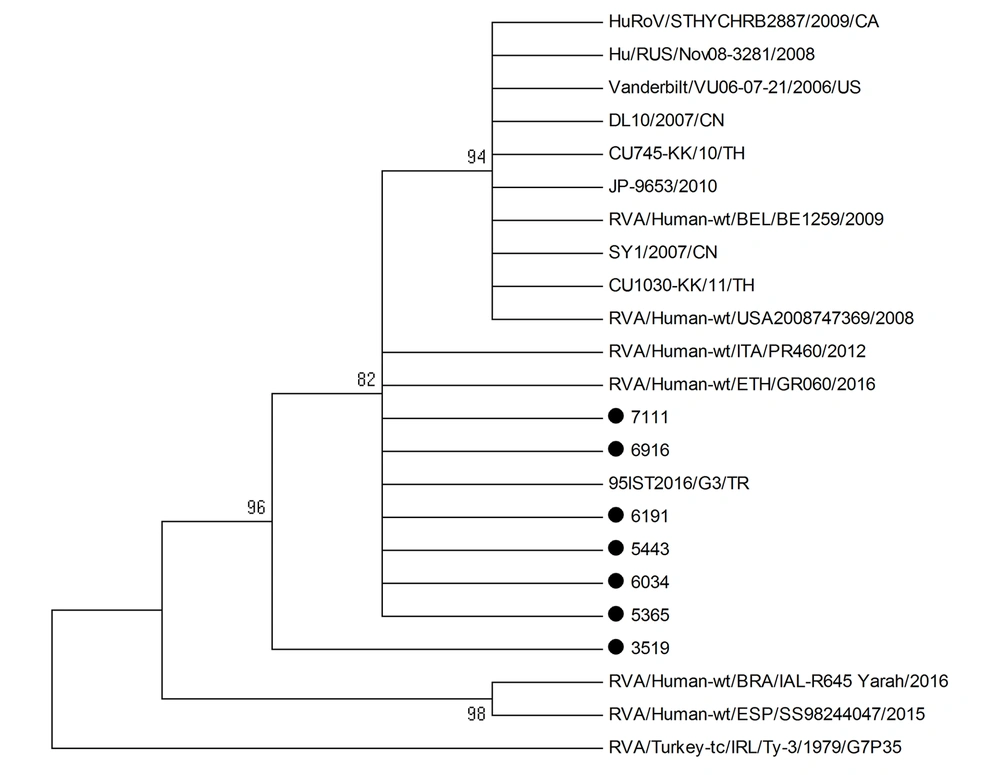
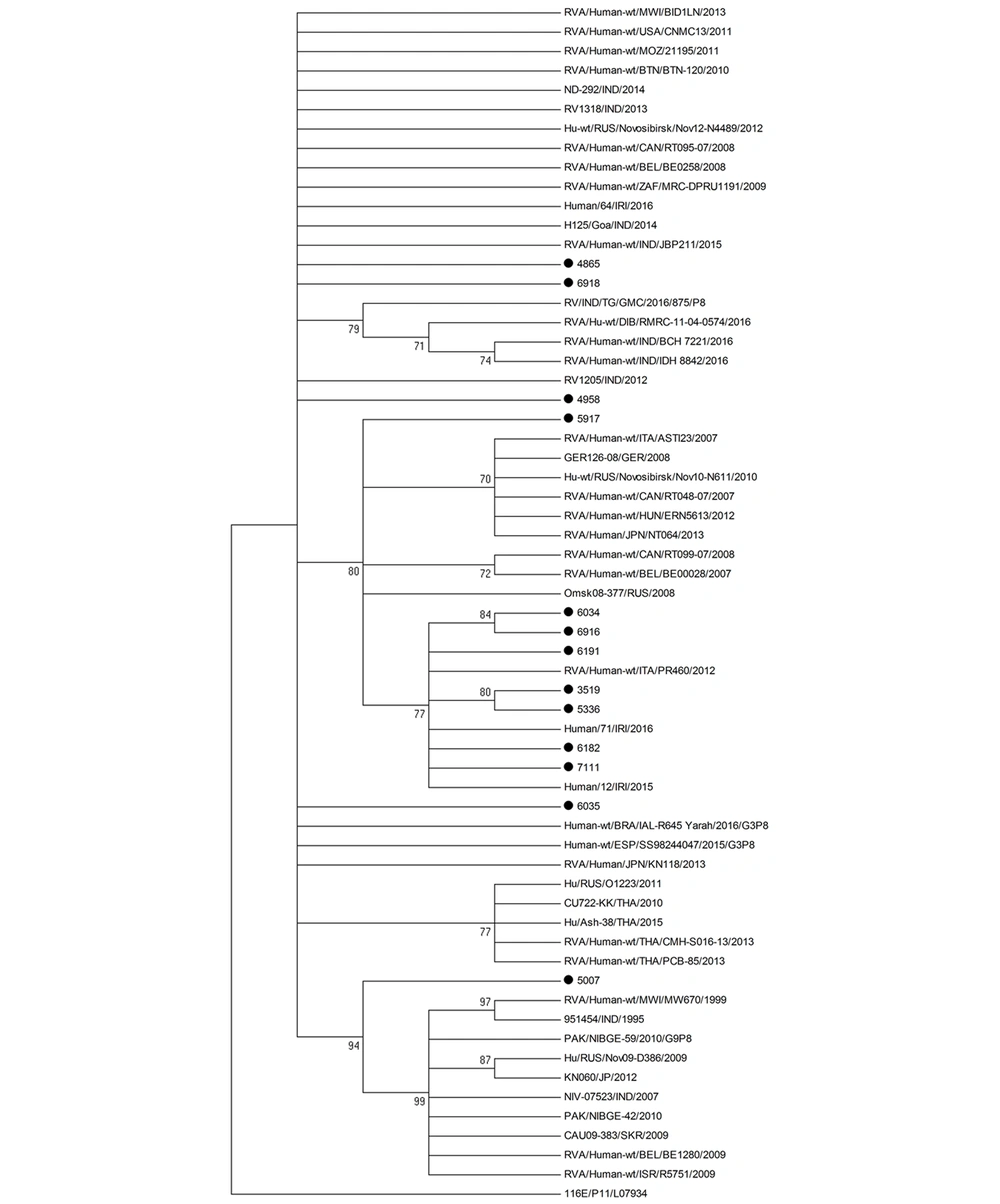
BALST analysis was used to compare the nucleotide sequences of rotavirus G3P[8] strains to those from global strains and those circulating during the same period. Only strains with nucleotide sequences of more than 600 bp were included in the analysis. The best hit results indicated high similarity with rotavirus sequences from different countries in Europe, North America, Africa, and Asia. However, our rotavirus strains failed to cluster with any strain in the phylogenetic tree (Figures 3 and 4). Besides, our rotavirus G3P[8] strains did not cluster with the equine-like rotavirus G3[P8] strain that emerged in 2013 as a dominant strain in different parts of the world, such as Australia, Germany, Hungary, Spain, Brazil, and in some countries within the Asia-Pacific region (21-25).
5. Discussion
In the present study, we analyzed partial nucleotide sequences of rotavirus VP4 and VP7 genes detected in stool specimens of children with severe diarrhea in the year preceding the introduction of RotaTeq vaccine in Kuwait. While G1P[8] was the most predominant genotype detected in more than 60% of children admitted to the same tertiary referral hospital in Kuwait about ten years ago (16), G3P[8] became the most common rotavirus type in 2016, accounting for about 50% of all strains detected. Based on the results of nucleotide sequence identities, the detected rotavirus strains were not vaccine-derived. G3P[8] is currently one of the six most common genotypes, causing 90% of severe diarrhea (26). Interestingly, the emergence of new genotypes such as G3P[8] has also been reported in India between 2012 and 2016, before the introduction of rotavirus vaccines (15).
All VP7 nucleotide sequences of G1, G3, G4, or G9 strains clustered in one genetic lineage, at least over the study period, suggesting the endemicity of the strains. The results of VP4 genotyping showing all P[8] strains but one formed one lineage are in agreement with this hypothesis. Moreover, our rotavirus G3P[8] strains failed to cluster with global strains and strains circulating during the same period. However, we could not compare G3P[8] strains that circulated during the year 2016 with those detected previously in Kuwait between 2005 and 2006. To confirm our results and exclude the introduction of different rotavirus lineages from the global population, the entire rotavirus genome should be analyzed.
Of note, all VP7 and VP4 nucleotide sequences of rotavirus G1P[8] type, and all VP4 nucleotide sequences of rotavirus G3P[8], G4P[8], or G9P[8] types clustered in lineages different from those to which belong Rotarix and RotaTeq vaccines. However, VP7 nucleotide sequences of rotavirus G3P[8] or G4P[8] type clustered within the same lineage as RotaTeq VP7. Rotavirus VP7 and VP4 are surface proteins that induce protective immunity (27), and both are represented in Rotarix and RotaTeq vaccines. Genetic diversity in VP7 and VP4 resulted in the emergence of lineages and sub-lineages among the rotavirus strains collected from different geographic locations (11). Rotavirus G9 strains were detected in three patients in two different combinations, G9P[8] and G9P[4]. Rotavirus G9P[8] strains were associated with diarrhea in different parts of the world (28) and were the predominant strains detected in neonates in India, between 1986 and 1993 (18).
The detection in our study of rotavirus G or P genotypes not included in the monovalent Rotarix vaccine or the polyvalent RotaTeq vaccine does not indicate vaccine inefficiency and a recommendation against vaccination. This may be explained by the presence in the vaccine of gene segments other than VP7 and VP4 that are involved in immunity as well (29). Indeed, the currently licensed vaccines, Rotarix and RotaTeq, were found to be effective against homotypic and heterotypic rotavirus strains, resulting in substantial declines in rotavirus-associated hospitalization in many countries (30-32). In addition, the G9P[4] emergence in Mexico was not related to vaccine pressure, vaccine failure, or failure to vaccinate, as the monovalent Rotarix vaccine was reported to be 94% effective against G9P[4] rotavirus-related hospitalization (33).
5.1. Conclusions
The findings of this study suggest a potential rise in the incidence of rotavirus G3P[8] in a major tertiary referral hospital in Kuwait in the prevaccine era. Also, our results highlight the circulation of genotypes and genetic lineages that are different from the strains included in the Rotarix or RotaTeq vaccine.