1. Background
Invasive fungal infections (IFI) have become a major concern for clinical management due to the increasing prevalence, emergence of new pathogenic species, and lack of efficient diagnostic tools (1). The widespread use of broad-spectrum antibiotics, increased use of invasive procedures (eg, intubation) and devices (eg, drains and catheters), and intensive care unit hospitalization are probably important contributing factors to the incidences of these infections (2).
Although Candida albicans is the most common yeast isolated from patients, other Candida spp. (ie, C. glabrata, C. guilliermondii, C. krusei, and C. tropicalis) have recently emerged as clinically important pathogens (3). Therefore, the finding and differentiation of the fungus at the species level are vital to accumulate antifungal healing, particularly for patients with IFI.
Phenotypic methods are still considered the gold standard in clinical laboratories. Morphological structures and generative constructions useful to identify divorced fungi might develop over days and weeks to be measurable in culture, and the estimation of these characteristics necessitates knowledge of mycology (4). Moreover, they cause a significant delay in diagnosis and treatment, which might lead to the failed management of fungal infections (4, 5).
During the past years, comparatively faster and more reliable techniques based on the molecular characterization of fungal species have been proposed to identify the most important fungal pathogens (6). Polymerase chain reaction (PCR)-constructed tests have progressively been used in laboratories to recognize fungal species.
A test with a quick completion time is possibly beneficial since it will not silence the occasionally life-preserving cure (7). In contrast, for the stimulation of the acceptance of the assay, the test should be easy, factually clarified, and economical in contrast to standard sequencing (8) with discrimination according to comparative amplification efficacy (9) or by probe-constructed assessments (10-12). Furthermore, the test should be strong to alterations initiated by pre-analytical features (13). Regarding the aforementioned issues, two paired approaches, including polymerase chain reaction-high resolution melt (PCR-HRM) and polymerase chain reaction-restriction fragment length morphism (PCR-RFLP) methods, were assessed in this study using the standard PCR and Sanger sequencing as the superior test. Both techniques, alone or in cooperation, might have the ability to substitute for present sequencing-centered methods.
2. Methods
2.1. Sample Assortment
This experimental study was conducted on 66 positive blood cultures collected from hospitalized patients referring to the medical mycology laboratory of a center for research and training in skin disease and leprosy, Tehran, Iran.
2.2. Laboratory Examination
This study utilized two types of culture medium, namely Sabouraud’s Dextrose Agar (Merck, Germany) and CHROMagar Candida (CHROMagar, France), and biphasic blood culture medium. The blood for the biphasic medium was collected directly from the patients, immediately injected into the bottles, and incubated at 37°C for 2 - 5 days. The culture-containing colonies were separated, and a colony of each culture was inoculated to CHROMagar Candida medium and incubated at 37°C for 24 - 48 hours.
2.3. DNA Extraction and PCR Sequencing
Deoxyribonucleic acid (DNA) was extracted from the blood samples using the DNG PLUS (Sinnacolon, Iran). The primers used for PCR amplification were ITS1-F (5'-TCC GTA GGT GAACCT GCG G-3') with an annealing temperature of 55.4°C and ITS4-R (5'-TCC TCC GCT TAT TGA TAT GC-3') (14). The PCR assay was carried out using 3 μL of the test sample (1 μL of each of primers, 10 μL of PCR Master Mix [Ampliqon, Denmark], and 9 μL of deionized distilled water).
The amplification was performed using the Eppendorf Thermal Cycler (Eppendorf SE, Germany) with the following program:
The early denaturation of DNA at 96°C for 5 minutes, 40 rounds involved a denaturation phase at 94°C for 30 seconds, an annealing phase at 58°C for 30 seconds, and a broadening step at 72°C for 30 seconds with a final broadening at 72°C for 15 minutes following the final round
Afterward, the amplicons were kept at 4°C until used for the RFLP technique. Negative controls were included for each running, and the presence of specific bands was examined by staining with DNA safe stain (Simbio, USA) and electrophoresis on 1.5% agarose gel. The PCR products were subjected to the sequence by Noorgeen Company (Iran).
2.4. Restriction Enzyme Analysis
All the PCR products were digested with MSPI enzyme to differentiate common yeasts. Briefly, 10 μL of the product was integrated with 1.5 μL of tango buffer and 0.5 U of the digestion enzyme. The final mixture with a volume of 12 μL was kept warm at 37°C for 2 hours. Then, the products were assessed using electrophoresis by DNA safe stain on 2% agarose gel.
2.5. Real-time PCR-HRM Analysis
Real-time PCR-HRM analysis was performed on a Corbett 6000 Real-time PCR System (Roche, Basel, Switzerland). Every tube contained 10 ng of DNA template and 0.2 µM of each primer. Real-time PCR was performed as follows:
Primary denaturation at 95°C for 10 minutes, later denaturation at 95°C for 10 seconds, annealing for 15 seconds with 0.5°C per run, from 65 to 55°C, and extension at 72°C for 10 seconds and 40 runes
For the PCR-HRM study, the HRM reactions (20 µL) contained 2X HRM Master Mix, 1 µL of each primer, and 2 µL DNA (15). Then, the amplicons were denatured at 95°C for 1 minute, renatured at 40°C for 1 minute, and a final step from 65 to 95°C (1°C each second). Real-time PCR was continuous for specimens with an overlapping-point run value higher than 28, optionally with an extra set of DNA decontamination using the QIAamp kit (USA) to remove bad amplification. The PCR-HRM curve analysis was performed using Corbett 6000 software using the default normalization for each.
3. Results
In culture examination, all of the blood samples, which were incubated in biphasic culture media, showed no sign of growth. The yeasts were then transferred to CHROMagar Candida for initial differentiation. In this culture media, the colonies of C. albicans shifted their color to green, C. glabrata to purple, C. tropicalis to blue, and C. krusei to pink purple. The samples, including C. albicans (n = 32), C. tropicalis (n = 11), C. glabrata (n = 7), C. krusei (n = 5), and C. parapsilosis (n = 11), were diagnosed.
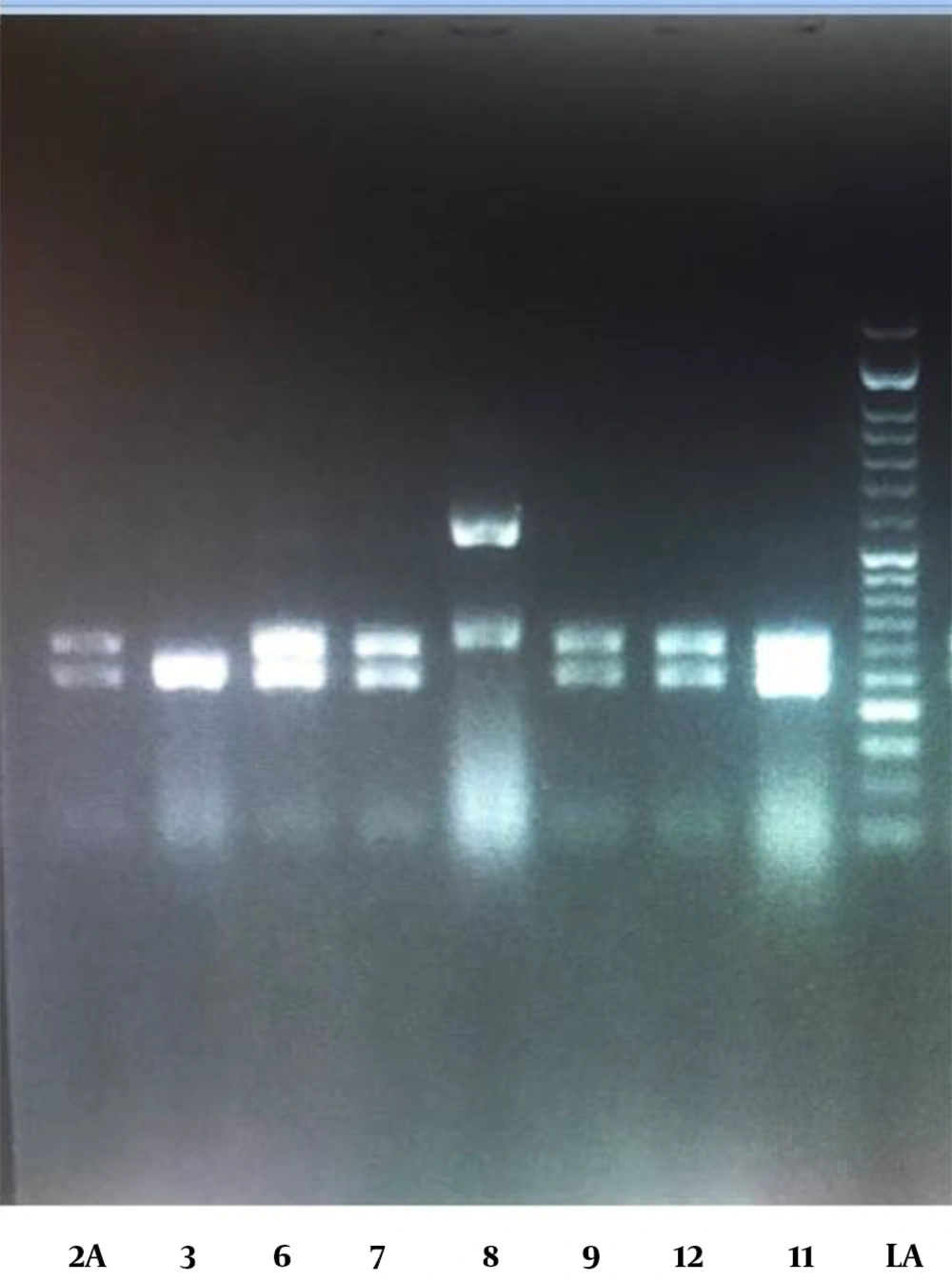
The PCR-RELP identification findings were also in absolute similarities with the Sanger sequencing results. The PCR-RFLP outcomes were defined according to the band pattern on the electrophoresis gel. Figure 1 illustrates the pattern of Candida spp. bands.
Polymerase chain reaction (PCR)-restriction fragment length polymorphism; agarose gel electrophoresis pattern of PCR products of Candida; lanes 2, 6, 7, 12, and 11: C. albicans due to their approximate 238 and 297 bp bands; sample 6 identified as C. krusei due to its approximate 249 and 261 bands; sample 8 identified as C. glabrata due to its approximate 314 and 557 bp bands
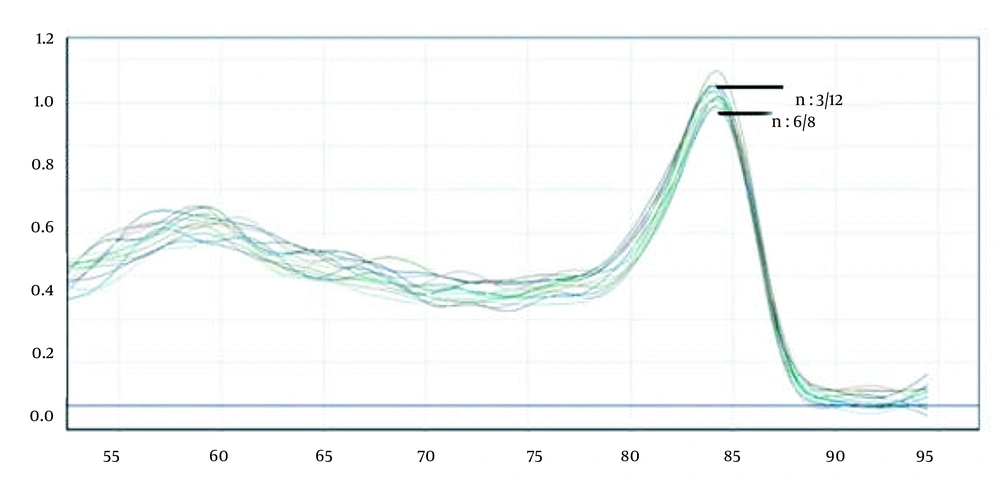
Based on Sanger sequencing, it was observed that there were only C. albicans from the C. albicans complex, and no samples of C. orthopsilosis and C. metapsilosis (ie, a member of C. parapsilosis complex) were noticed in this study. Further analysis, which could be assigned to all groups, was performed on Corbett 6000 software (Figure 2). The samples diagnosed using PCR-HRM were C. albicans (n = 31), C. africana (n = 1), C. tropicalis (n = 11), C. glabrata (n = 7), C. krusei (n = 5), C. parapsilosis (n = 10), and C. orthopsilosis (n = 1). The close curve changes were not exhibited for isolated 3/12 (ie, C. albicans/C. africana) and 6/8 (ie, C. parapsilosis/C. orthopsilosis).
Furthermore, there were samples that were detected negative in the culture results of microscopic analysis; however, they were detected positive by PCR-RFLP and PCR-HRM methods. This could be due to the increased sensitivity of PCR-RFLP and PCR-HRM approaches to detect very low amounts of fungal DNA in suspected samples.
4. Discussion
Conventional Sanger sequencing has been considered the “superior". For numerous clinical purposes, this method is preferred and remains valid since the precise sequence change might be unidentified, and the finding and characterization of novel species are of diagnostic importance. In this study, two alternative methods were advanced for the recognition of clinically valuable Candida spp. When melting curves overlap among some species, it can be stated that the pun fungal primer in the PCR-HRM method fails to differentiate the majority of the closely related species. Therefore, RFLP offered a supportive analytic benefit with specific sensitivity in distinguishing these species (inadequate restriction digestions would be more sensitively detected by PCR-HRM analysis). In the present investigation, no disagreements were observed between PCR-HRM- and RFLP-constructed tests except for C. albilcans and C. parapsilosis complexes.
After comparing the three procedures, Sanger sequencing remains the standard reference technique due to its accuracy, capability to identify each novel species, and comparative strength to record DNA quantity and quality. Additional optimizations, containing cumulative the number of PCR sets (16), might be due to a decrease in the speculative restriction of a genomic copy (17); nevertheless, since a medical laboratory performing analytical evaluating, it might be superior to balance within the reasonable use of DNA, the acceptable amount of PCR rounds, and standardizing laboratory procedures to escape extreme pipetting and dilution phases. Concerning the aforementioned issues, PCR-HRM is considered less robust, compared to Sanger sequencing and PCR-RFLP, which rely only on the existence of acceptable amplicon and digestion enzymes instead of similar amplification efficiency (18).
It should be noticed that PCR-HRM and PCR-RFLP have a more economic component than sequencing and an improvement time, one-third to one-fourth of that of sequencing. Moreover, the identification with these methods does not need a physical examination of sequencing evidence, and their explanation can be measured moderately forthright. The real growth in cost-effectiveness and usefulness might differ depending on the local reagent and labor costs; however, it is estimated that the acceptance of both substitute approaches will result in reduced cost and time, compared to the Sanger sequencing.
The limitation of the current investigation was the relatively small number of samples (n = 60), which disallowed broad justification of the methods.
4.1. Conclusions
In conclusion, given the number of studies performed on the comparison of sensitivity and specificity of phenotypic and genotypic methods to diagnose and identify invasive fungal pathogens and the findings of this study, it could be stated that the complementary real-time PCR-HRM and PCR-RFLP methods were effectively advanced as substitutes for conventional Sanger sequencing for economic identification. However, supplementary evaluations and confirming studies should be carried out with a broad range of samples to standardize this method for routine application in medical laboratories.