1. Context
In December 2019, a new virus was introduced as the cause of upper and lower respiratory tract infections in Wuhan, Hubei Province, China (1). Genetic studies revealed that the cause of this disease was a new virus called Coronavirus disease 2019) COVID-19) (1). The disease spread rapidly and was transmitted like a contagious disease throughout China, and then it gradually spread in other parts of the world.
Iran was also among the first countries involved in the COVID-19 pandemic, and the first official case of COVID-19 in Iran was reported on 2020.02.20 (2). By then, 738,322 SARS-CoV-2 positive patients and 40,582 deaths were detected in Iran.
The disease can be characterized as asymptomatic and fatal (1, 3-5). This is while the real frequency of the patients has remained undetected, which is a challenging epidemiological problem (6). Accordingly, diagnosing asymptomatic carriers is vital to adopt effective measures in high-risk environments such as hospitals (7).
The COVID-19 virus infects the lungs more than other organs because the virus enters host cells via the ACE2 enzyme, which is abundant in type II alveolar cells of the lungs (8). As the disease progresses, respiratory failure may occur, and eventually, it can lead to death. Coronavirus can affect gastrointestinal organs regarding the ACE2 expression on gastric glandular cells, rectal epithelium, duodenal, small intestinal enterocytes, and endothelial cells (9, 10). In this regard, the exact incubation period, however, is unknown, and it is assumed that it lasts about 2 - 14 days after exposure to the virus, with cases occurring during five days after exposure being as the most frequent ones.
The rapid and accurate detection of the SARS-CoV-2 virus plays a vital role in selecting timely treatments, saving lives, and preventing the spread of the disease. One of the crucial points in controlling epidemics is performing timely diagnostic tests, isolating patients, and reviewing their close contacts. Diagnostic tests can play a critical role in identifying and isolating patients and preventing the spread of the pandemic. The success of many countries in controlling this pandemic has been the result of their access to relevant tests and their rapid response to infection control. One of the indicators indicating the accountability of health systems in this pandemic is the number of tests performed per one million persons. The statistics have reported 115,000 tests/persons (in million) in Iceland, and about 20,000 tests/persons (in million) in Germany, Spain, and Italy; however, it is about 10,000 and about 4,000 tests/persons (in million) in the United States and Iran, respectively. The smallest rates are < 500 for African countries. The urgent need is to perform the test for hospitalized patients with suspicious symptoms and then health service providers with symptoms. Testing individuals with mild symptoms or even asymptomatic individuals having a history of contact can help control the disease. Here are some criteria to be considered regarding decisions on how to choose a test and how to perform it: (1) need for clinical diagnosis; (2) speed of test; (3) number of samples to be collected for an affordable experiment; (4) indications of current infection or just a previous infection; (5) time when the test results are provided; (6) number of samples to be tested simultaneously; (7) facilities required to perform the test; (8) possibility of performing the test at a patient's primary visit site; and (9) provision of primary screening or definitive diagnosis?
In line with our recent studies on COVID-19 (11-13) and to help diagnose SARS-CoV-2, this study summarizes the methods used to identify coronavirus nucleic acid and examines the effectiveness of coronavirus nucleic acid detection kits as well as the performance of other diagnostic techniques. It is hoped that the findings would help researchers and physicians develop better techniques for the timely and effective diagnosis of coronavirus infections.
2. Methods
2.1. Data Sources
The collected articles were extracted by a systematic search of the Embase, Google Scholar, MEDLINE, Web of Science, Scopus, and PubMed databases as well as the references of all relevant articles in English published during 2019 - 2020 using keywords related to COVID-19, detection kits, and respiratory failure and proceedings from relevant conferences and congresses.
2.2. Search Strategy
All papers were searched using the following keywords and their combinations: “COVID-19” , “SARS-CoV-2” “detection kits,” “respiratory failure,” “Laboratory tests,” “Laborartory diagnosis,” “Serology testing,” “RT-PCR,” “Nasopharyngeal sampling,” “Oropharyngeal sampling,” “Saliva,” “Urine,” “Feces,” “Lower respiratory tract,” “Upper respiratory tract,” “Bronchoalveolar washing,” “Sputum,” “IgM and IgG antibody,” and “Antigenic tests.”
2.3. Research Objective
The present study summarizes the methods used to identify coronavirus nucleic acid and examines the effectiveness of coronavirus nucleic acid detection kits for different samples as well as the performance of other diagnostic techniques.
2.4. Data Extraction
The collected papers were analyzed in terms of the place and date of publication, age of participants, resources, control groups, project design, human types, outcomes, measurement instrumentation, and conclusions. The authors examined the studies independently and detected the aforementioned information.
3. Results
The results of previous studies are classified into several categories, including all diagnosis methods of the COVID-19 disease. Table 1 summarizes the findings.
| Authors | Study Subjects | Objectives | Outcomes |
|---|---|---|---|
| Li et al. 2020 (14) | Blood cases from 397 PCR confirmed COVID‐19 patients and 128 negative patients | To evaluate the clinical detection sensitivity and specificity of quick tests in identifying COVID-19. | The outcomes revealed that the sensitivity of this technique was 88.66%, and its specificity was 90.63%. |
| Tosato et al. 2020 (15) | Blood samples from 133 asymptomatic health workers | To examine the levels of IgM and IgG antibodies by MAGLUMI 2019-nCoV IgM/IgG kit. | The sensitivity and specificity of this kit were 78.65 and 97.50% for IgM, 91.21 and 97.33% for IgG, and 89.89 - 95.6% and 96.5 - 96.0% for IgM + IgG. |
| Yongchen et al. 2020 (16) | Blood samples from 21 persons infected with COVID-19 | To study the relationship between molecular methods and serological techniques. | The serological technique is a method supplementary to molecular assay in those symptomatic SARS-CoV-2 cases. |
| Xie et al. 2020 (17) | 19 suspect samples | To survey laboratory and radiological features of 19 patients using RT-PCR test on blood, anal, oropharyngeal, and urine samples. | The SARS-CoV-2 virus was diagnosed in nine patients using oropharyngeal samples, and this virus was identified in eight out of these nine patients by anal swabs. No positive result was noticed in blood and urine samples. |
| Pan et al. 2020 (18) | 23 confirmed cases with COVID-19 infection | Effects of heat on RNA related to COVID-19 virus inactivation. | Thermal inactivation had a negative effect on RT-PCR efficiency. |
| Lou et al. 2020 (19) | 80 cases with PCR-confirmed SARS-CoV-2 | To examine the serological response to the COVID-19 infection by ELISA assay. | IgG, total antibody, and IgM were 93.8, 98.8, and 93.8%, respectively. |
| Long et al. 2020 (20) | 285 cases | To determine the antibody response to the COVID-19 virus. | Serological method may be effective in the detection of suspected cases with negative RT-PCR outcomes. |
| Chan et al. 2020 (21) | 273 clinical samples from 15 COVID-19 patients | To determine three methods [(RdRp)/helicase (Hel), spike (S), and nucleocapsid (N)]. | The detection of COVID-19 by the (RdRp) / helicase (Hel) method did not interact with other respiratory viruses and human coronaviruses. The COVID-19-RdRp/Hel assay was significantly more sensitive than the RdRp-P2 247 assay in the detection of SARS-CoV-2 RNA using nasopharyngeal aspirates/swabs or throat swabs 248 (P = 0.043), saliva (P < 0.001), and plasma (P = 0.001) specimens. |
| Xiong et al. 2020 (22) | Human specimens (n = 46). | To investigate the efficiency of four methods (Daan, Sansure, Hybribio, and Bioperfectus) in the diagnosis of COVID-19. | The limit of detection (LOD) of three methods (Daan, Sansure, and Hybribio) was 3,000 copies per milliliter, while LOD of Bioperfectus was 4000 copies per milliliter. Moreover, the sensitivity and specificity of the three methods were 100%; however, the specificity of the Bioperfectus method was 100%, and its sensitivity was 81.25%. |
| Lin et al. 2020 (23) | 52 patients with suspected SARS-Cov-2 | To study the throat and sputum samples for the diagnosis of viral nucleic acid by the RT-PCR. | The positive rates of COVID-19 from sputum samples was76.9%, and the positive rates of throat swabs was 44.2%. |
3.1. Quick Diagnostic Tests of COVID-19
Rapid tests qualitatively detect IgG and IgM antibodies in human blood, serum, and plasma using lateral chromatography immunosuppression. The IgM-IgG combination method is more sensitive than the IgM or IgG single tests. This method can be used to rapidly screen SARS-CoV-2 carriers, symptomatic or asymptomatic, in hospitals, clinics, and laboratories (14). The covid-19 rapid test kit is presented in Figure 1.
COVID-19 rapid test kit (24)
Li et al. established a rapid IgG‐IgM combined antibody test using lateral flow immune assay methods and showed that the sensitivity of this technique was 88.66%, and that its specificity was 90.63% (14).
Pan et al. also used serological colloidal gold-based immunochromatographic (ICG) strip assay to diagnose SARS-CoV-2. They documented that the sensitivity of this method was 11.1% in the early stage (1 - 7 days after onset), 92.9% in intermediate stage (8 - 14 days after onset), and 96.8% in the late stage (more than 15 days) in the RT-PCR positive patients group. In the RT-PCR negative group, the detection rate of this serologic method was 43.6% (18).
Suhandynata et al. also studied the association between IgM and IgG and molecular techniques levels. They expressed that serological techniques are considered a supplementary method to support the RT-PCR technique in detecting asymptomatic patients (25).
3.2. Diagnostic Methods Relying on Molecular and Nucleic Acid [Reverse Transcription-Polymerase Chain Reaction (RT-PCR)]
The RNA of the COVID-19 virus is detected by a standard test method (RT-PCR). The results are available to physicians during a few hours to two days. Various studies have shown that this diagnostic method has a specificity of about 70% and a sensitivity of 95%.
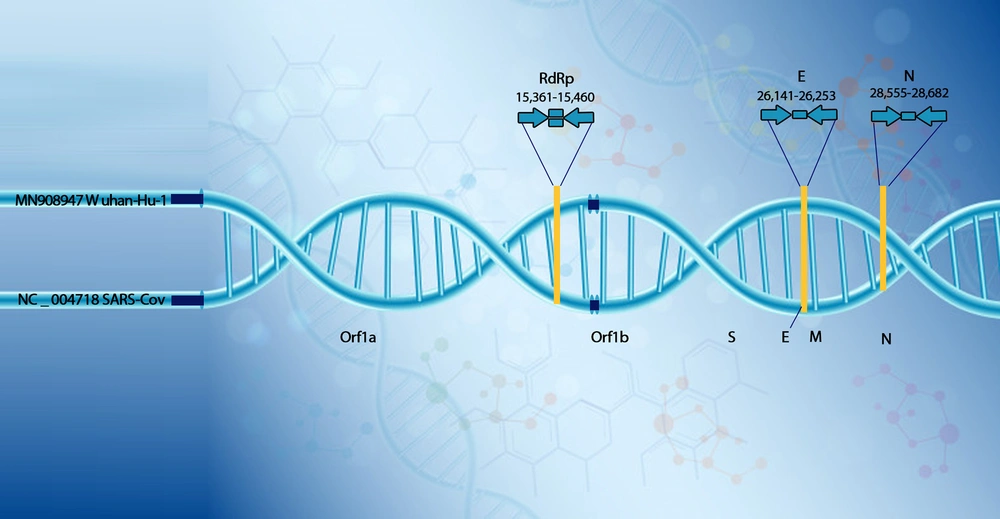
This test is performed on the COVID-19 open reading frame 1ab (ORF1ab) and nucleocapsid protein (NP) genes fragments (26) by valid kits under certain conditions (50°C for 15 min, 95°C for 3 min, followed by 45 cycles of 95°C for 15 s and 60°C for 30 s) (27). According to previous studies, when two fragments (ORF1ab, NP) were positive using RT-PCR, it was considered as a positive laboratory case (26). Figure 2 represents the genome of SARS-CoV-2.
Suitable locations of amplicon targets on SARS coronavirus and COVID-19 genome (E, envelope protein gene; M, membrane protein gene; N, nucleocapsid protein gene; ORF, open reading frame; RdRp, RNA-dependent RNA polymerase gene; S, spike protein gene; Numbers below amplicons are genome positions according to SARS-CoV, GenBank, and NC_004718) (28).
The method and the location of sampling have a significant impact on the sensitivity of this experiment. This test is usually performed on respiratory samples collected by the nasopharyngeal swab; however, samples obtained using a nasal swab or sputum sample may be used. Various studies are being conducted to diagnose COVID-19 by RT-PCR, and researchers have been examining different samples with new methods and new kits to help individuals with the correct and rapid diagnosis of COVID-19 disease.
In a study, Chan et al. examined the RT-PCR method with a target gene different from the aforementioned genes to diagnose the COVID-19 virus. They studied the performance of (RdRp)/helicase (Hel) and concluded that the detection of COVID-19 by the (RdRp)/helicase (Hel) did not interact with other respiratory viruses and human-pathogenic coronaviruses in clinical samples. They showed that this highly sensitive and specific assay might promote human health by improving the COVID-19 laboratory diagnosis (21).
In another study, Xiong et al. researched the efficiency of four reagents (Daan, Sansure, Hybribio, and Bioperfectus) commonly used for the RT-PCR of COVID-19. They reported that the sensitivity of these four reagents was significantly lower than what their manufacturers had declared. They also showed that the limit of detection (LOD) of three methods (namely Daan, Sansure, and Hybribio) was 3,000 copies per milliliter, while the LOD of Bioperfectus was 4000 copies per milliliter. Furthermore, they claimed that the sensitivity and specificity of the three methods (namely Daan, Sansure, and Hybribio) were 100%; however, the specificity of the Bioperfectus was 100%, and its sensitivity was 81.25%. Accordingly, the four methods had high specificity; however, Bioperfectus may be dangerous for detecting low-virus specimens (22).
Zhao et al. developed a viral RNA extraction method based on magnetic nanoparticles for the detection of COVID-19. In this study, they reported the synthesis of polyamine ester with carboxylic compounds-coated magnetic nanoparticles (PCMNPs) and used this combination to develop a PCMNPs-based viral RNA extraction method for the detection of SARS-CoV-2. According to the researchers, this method combines the steps of lysis and binding in one step, and pcMNPs-RNA complexes can be directly included into the following RT-PCR reactions. Accordingly, due to the excellent performance and simplicity of this new extraction method, it can help the earlier identification of the COVID-19 disease than the usual RT-PCR method (29).
In a study, Broughton et al. developed and confirmed a quick, inexpensive, and precise CRISPR-Cas12 method based on lateral flow to detect COVID-19 virus. This method was first applied by RT-LAMP from nasopharyngeal or oropharyngeal swabs, followed by the Cas12 detection of COVID-19 sequences. It is a qualitative method with less analytical sensitivity than RT-PCR; however, its turnaround time lasts about 45 minutes (30).
A quick and low-cost way to identify the COVID-19 virus has been developed to detect the virus in less than 30 minutes. This method is called Reverse Transcription loop-mediated isothermal amplification (RT-LAMP), and several studies have examined this method. They concluded that RT-LAMP method could be used for screening individuals exposed to the COVID-19 virus (31). In another study, Zhang et al. used the RT-LAMP method to diagnose SARS-CoV-2 and showed that this sensitive and simple technique provided an opportunity for the identification of COVID-19 virus without the need for complex infrastructure. Moreover, they compared the accuracy of RT-LAMP and RT-PCR, as shown in Table 2 (32).
Abbreviations: +, positive reaction; -, negative reaction; N.A., not detected.
a No template control.
b Plasmid DNA (standard control) for RT-PCR kit.
The type of sample is also critical in detecting the COVID-19 virus. In a study, Lin et al. showed that the diagnosis rate of COVID-19 from sputum samples using the RT-PCR method was significantly higher than that of throat samples (P = 0.001). Accordingly, the results of their study can help the selection of samples and enhance the accuracy of diagnosis (23).
Long et al. examined the findings of CT and rRT-PCR techniques to diagnose COVID-19 and showed that the preliminary outcomes of rRT-PCR might be false negative. They recommend that individuals with normal CT results and negative rRT-PCR outcomes be isolated, and rRT-PCR be repeated. Moreover, the preliminary initial CT had a sensitivity of 97.2%, whereas the sensitivity of the preliminary initial rRT-PCR sensitivity was 83.3% (20).
Garg et al. examined seven commercial RT‐PCR kits to diagnose SARS‐CoV‐2: (1) TRUPCR SARS‐CoV‐2 Kit (Black Bio); (2) TaqPath RT‐PCR COVID‐19 Kit (Thermo Fisher Scientific); (3) Allplex 2019‐nCOV Assay (Seegene); (4) Patho detect COVID‐19 PCR kit (My Lab); (5) LabGun COVID‐19 RT‐PCR Kit (Lab Genomics, Korea); (6) Fosun COVID‐19 RT‐PCR detection kit (Fosun Ltd.); (7) real‐time fluorescent RT‐PCR kit for SARS CoV‐2 (BGI). They reported that all the seven RT-PCR kits could be used for the molecular detection of COVID-19; however, LabGun kit, Patho kit, and Fosun detection kits could detect 85, 75 and 90% of weakly positive samples, respectively (33).
Furthermore, Hur et al. tested four commercial RT‐PCR kits (namely Allplex 2019-nCoV Real-time PCR (Seegene, Seoul, Korea), PowerChek 2019-nCoV (KogeneBiotech, Seoul), Real-Q 2019-nCoV Real-Time Detection (BioSewoom, Seoul), and StandardM nCoV Detection (SD BIOSENSOR, Osong, Korea) to detect COVID‐19. They indicated that none of the four kits had cross-reactivity with other respiratory viruses. Furthermore, it was found that the four kits were appropriate for identifying and following the COVID-19 test (34). Furthermore, other studies have examined the RT‐PCR kits and achieved relevant findings (35, 36).
Liu et al. used stool samples of 69 patients to identify the COVID-19 virus and concluded that the existing techniques are not comprehensive for the detection of the virus at oral/nasopharyngeal swabs since the live COVID-19 may be present in the stool samples, while oropharyngeal or nasopharyngeal samples are negative (37).
In a study, Zhang et al. also used oral samples and anal samples and noticed more positives in anal swabs than oral samples, indicating that the COVID-19 virus was a novel virus and might be transmitted by various routes (38).
Moreover, Xie et al. examined the laboratory and radiological features of 19 patients by the RT-PCR test using blood, anal, oropharyngeal, and urine samples. In their study, the SARS-CoV-2 virus was diagnosed in nine patients using oropharyngeal samples, and this virus was identified in eight of the patients by anal swabs. No positive result was noticed in blood and urine samples. Accordingly, both molecular and radiographic methods must be used to detect the SARS-CoV-2 virus (17). Wu et al. used several samples (nasopharyngeal swab, sputum, blood, anal swabs, and feces) and the RT-PCR test for the diagnosis of the SARS-CoV-2 virus in 132 patients. They found that the sensitivity of samples was as follows: nasopharyngeal swab (38.13%), the sputum (48.68%), blood (3.03%), anal swabs (10.00%), and feces (9.83%) (39).
In an interesting study, Pan et al. investigated the effects of heat on the RNA of COVID-19 virus inactivation and examined the false-negative results of the RT-PCR method. They documented that thermal inactivation had a negative effect on RT-PCR efficiency in the diagnosis of COVID-19 virus (40).
3.3. Serological Evaluation of Antibody Response to COVID-19 in Blood
Although molecular techniques have become common approaches in detecting infectious illnesses, serological methods have been, for many decades, considered as the main diagnostic tools for many diseases, including HIV and hepatitis B and C (41).
Serological tests play a vital role in diagnosing asymptomatic cases (7, 42, 43). The serosurvey has two key advantages as it can be considered as a tool (1) to determine local transmission factors and risk factors of infection (42, 43); and (2) it to assess protective immunity (42).
Although serological methods play an critical role in the diagnosis of infectious sicknesses, these techniques have limitations. Significant variation in the kinetics and value of the serological response, particularly in the early stages of the disease, could lead to false negative outcomes. Furthermore, a wide range of agents can affect the specificity and sensitivity of serological methods. The results of false positive, which can incredibly happen with IgM, causes errors in the accurate interpretation of the findings. Despite such shortcomings, repeating serological methods may be helpful under acute conditions (41).
Serological tests are often considered a supplement to the molecular method (RT-PCR) in diagnosing COVID-19 infection (44). They can be beneficial for the quick diagnosis of patients and thus the detection of the next chains of emergence to identify close contacts and recommend quarantine (45).
In this regard, various studies have been conducted. In our previous study measuring the IgG level of COVID-19 using the ELISA technique, it was revealed that there was anti-SARS-CoV-2 IgG in the blood of 29 patients, and that a majority of individuals did not have anti-SARS-CoV-2 IgG. Moreover, 320 patients had at least one clinical sign within the last six months. Moreover, among PCR-positive persons, nine cases had anti-SARS-CoV-2 IgG, compared to seven case of positive CT. Accordingly, education and technical advice on COVID-19 is necessary (12).
Studies examining the prevalence of IgG of COVID-19 using the ELISA technique in clinical environments have reached inconsistent results. For example, in a hospital in Fujisawa, the serum prevalence of this antibody among medical staff was 0.74% (46); however, its prevalence was 17% in another study at Nanjing Drum Tower Hospital in China (7).
Considering the ELISA technique, other studies have indicated that the sero-prevalence of IgG in SARS-CoV-2 among medical personnel was 11% (Belgium) (47), 1.2% (Germany) (48), 0.83% (USA) (49), 2.0% (China) (26), 1.7% (Italy) (27), 2.7% (Denmark) (50), 7.4% (Italy) (51), and 4.9% (Michigan, USA) (52).
Amanat et al. developed a serological technique for the diagnosis of COVID-19 in humans. They reported that this specific and sensitive enzyme-linked immunosorbent assays (ELISA) method facilitated screening and detecting COVID-19 by human serum/plasma two days after the onset of symptoms (53).
Moreover, Zhang et al. examined the detection positive rate of the SARS-CoV-2 virus using ELISA-based diagnostic methods. They revealed that IgM and IgG antibodies were somewhat negligible or unrecognizable on day 0 (first day of sampling); however, on day 5, an increase was noticed in antibodies in almost all patients, implying that serological techniques can contribute to confirming COVID-19 infection (38).
In their study, Xiang et al. examined the diagnosis of COVID-19 disease using the IgM and IgG antibodies by the ELISA technique. They concluded that in SARS-CoV-2 patients, positive predictive value (PPV), sensitivity, negative predictive value (NPV), and specificity were100, 77.3, 80.0, and 100% in terms of IgM, and 94.8, 83.3, 83.8, and 95.0% in terms of IgG, respectively. In patients with suspected SARS-CoV-2 positive predictive value (PPV), sensitivity, negative predictive value (NPV) and specificity were 100, 87.5, 95.2 and 100% in terms of IgM, and 85.0, 70.8, 89.1, and 96.6% in terms of IgG (54). In this study, seroconversion was noticed four days after the emergence of symptom.
Lou et al. examined the serological response to the COVID-19 infection by ELISA assay. They showed that the detectable levels of total antibody, IgM, and IgG were observed after 9, 10, and 12 days from symptoms beginning with 98.8, 93.8, and 93.8% seroconversion rates, respectively. This method can be a supplement to the RT-PCR technique (19).
Long et al. analyzed the antibody response to the COVID-19 virus in 285 cases by magnetic chemiluminescence enzyme immunoassay (MCLIA). Serum changes related to IgG and IgM with this method was 13 days after the onset of symptoms. This study concluded that the serological method might effectively identify suspected cases with negative RT-PCR outcomes as well as asymptomatic patients (55).
In this regard, Tosato et al. in Italy examined IgM and IgG antibodies in 133 asymptomatic health workers using the MAGLUMI 2019-nCoV IgM/IgG kit. In their study, the sensitivity and specificity of this kit were 78.65 and 97.50% for IgM, 91.21 and 97.33% for IgG, and 89.89 - 95.6% and 96.5 - 96.0% for IgM + IgG. They could use this method to screen health workers (15).
3.4. Antigenic Detection Methods
In addition to the above methods, there are antigenic detection methods for influenza and other respiratory viruses. In addition to being rapid, these methods were also cheap techniques but had no appropriate sensitivity and specificity (56-60). Unlike molecular techniques the antigen methods have no amplifcationsteps, and like PCR-based techniques the antigen tests directly identify viral ingredients (such as protein N released, protein M and glycoprotein S) or virus, and both of them are usefull for active viral infection stage (61). Antigenic methods can be used for the rapid detection of LFA strips with better sensitivity in ELISA.
In a study, Diao et al. established a fluorescent immunochromatographic LFA technique to detect the nucleocapsid (N) protein of SARS-CoV-2 (62). Though ELISA and LFA methods are well-known and used to detect COVID-19 infection, antigen detection techniques are highly required, so an antigen detection test for SARS-COV-2, known as the "Sofa 2 SARS antigen test kit" (U.S. Food and Drug Administration) was developed. This diagnostic kit is based on sandwich-type immunofluorescence strip technology and is used to identify N proteins of both SARS-CoV and SARS-CoV-2. Moreover, this kit uses nasopharyngeal and nasal swab samples and is only utilized by medical experts. This kit has 80% clinical sensitivity, while its specificity was 100% among 47 positive and 96 negative clinical samples (U.S. Food and Drug Administration).
After the failures of several antibody kits (Cohen, 2020a; Royal Statistical Society Covid-19 Task Force, 2020), WHO and FDA warned scientific and medical communities on the use of the rapid commercial tests for SARS-CoV-2 detection (U.S. Food and Drug Administration, 2020c). According to WHO, "with the limited data now available, WHO does not currently recommend using antigen-detecting rapid diagnostic tests for patient care, although research into their performance and potential diagnostic utility is highly encouraged" (WHO, 2020b).
Several studies have been performed to detect the COVID-19 virus by antigen detection. Lambert-Niclot et al. used this method by nasopharyngeal swab to detect the virus and showed that this technique had the specificity of 100% and the sensitivity of 50% (63). In another study, Mertens et al. used SARS-CoV-2 Ag respi-strip diagnostic technique and indicated that this method had the sensitivity of 57.6% and the specificity of 99.5%. They claimed that the method could play a complementary role for molecular methods (64).
4. Discussion
Since December 2019, there have been some cases of pneumonia in China with clinical symptoms highly similar to viral pneumonia (14). Due to the rapid spread of this disease worldwide, which threatened people's lives, researchers have provided various methods to diagnose this disease and achieved different results. In this study, we summarized and compared these methods. To sum up, there are different methods to diagnose the SARS-CoV-2 disease.
In the pre-analytical step, it is necessary to collect the appropriate respiratory samples at an appropriate time from the appropriate anatomical location for the rapid and accurate detection of SARS-CoV-2 disease. Moreover, appropriate actions are necessary to keep laboratory personnel safe during the production of valid results. In the analytical step, the RT-PCR test is a crucial method to detect COVID-19, while antibody-based assays are complementary devices. In the post-analytical step, results should be cautiously interpreted using serological and molecular outcomes (56).
Moreover, rapid tests are used to detect IgG and IgM antibodies in COVID-19 disease (14, 18). The reverse transcription-polymerase chain reaction (RT-PCR) is a standard gold tool for the detection of the COVID-19 virus. This method has been performed on different samples (throat, sputum, blood, anal, oropharyngeal, and urine). This technique can be performed in three ways [(RdRp)/helicase (Hel), spike (S), and nucleocapsid (N)], among which the (RdRp) / helicase (Hel) method is the most sensitive and the most specific (21). The project studied the efficiency of four methods (Daan, Sansure, Hybribio, and Bioperfectus) to detect SARS-CoV-2.
In another study, the CT and rRT-PCR techniques were tested to diagnose COVID-19. Moreover, the preliminary initial CT had a sensitivity of 97.2%, whereas the sensitivity of the preliminary initial rRT-PCR sensitivity was 83.3% (20).
Moreover, in a project was examined seven commercial RT‐PCR kits to identify SARS‐CoV‐2, and it was found that all seven RT-PCR kits could be used for the molecular detection of COVID-19. However, LabGun kit, Patho kit, and Fosun detection kits could detect 85, 75, and 90% of weakly positive samples, respectively (34).
Hur et al. tested four commercial RT‐PCR kits to detect COVID‐19 and indicated that none of the four kits had cross-reactivity with other respiratory viruses. Moreover, it was found that four kits were effective in identifying and following the COVID-19 test (34). Furthermore, other studies examined the RT‐PCR kits and achieved promising results (35, 36).
The sensitivity and specificity of the three methods (Daan, Sansure, and Hybribio) were 100%; however, the specificity of the Bioperfectus method was 100%, and its sensitivity was 81.25% (22). Using a viral RNA extraction method based on magnetic nanoparticles can decrease the return time and operational equipment in the molecular detection of COVID-19 (29). Several studies have been conducted on the RT-LAMP method, and it has been revealed that this technique detects the COVID-19 virus in less than 30 minutes and is an appropriate tool for the identification of disease (23, 31, 32). ELISA-based diagnostic methods are being commercialized to detect IgM and IgG antibodies against the SARS-CoV-2 virus, which are often produced against the nucleocapsid virus. These kits are being evaluated for their diagnostic values. The kits are mainly used to diagnose infection within two weeks. One of the main applications of these kits is to study the epidemiology of COVID-19 disease. In this regard, researchers conducted various studies to diagnose the SARS-CoV-2 virus using ELISA technique and determined that the ELISA method could identify the COVID-19 virus by measuring IgM and IgG antibodies (19, 38, 53-55).
In our previous study measuring the IgG level of COVID-19 using the ELISA technique, it was revealed that there was anti-SARS-CoV-2 IgG in the blood of 29 patients, and that a majority of individuals did not have anti-SARS-CoV-2 IgG. Accordingly, education and technical advice on COVID-19 is necessary (12).
In a hospital in Fujisawa, the serum prevalence of this antibody among medical staff was 0.74% (46); however, its prevalence was 17% in another study at Nanjing Drum Tower Hospital in China (7).
Considering the ELISA technique, other studies have indicated that the sero-prevalence of IgG in SARS-CoV-2 among medical personnel was 11% (Belgium) (47), 1.2% (Germany) (48), 0.83% (USA) (49), 2.0% (China) (26), 1.7% (Italy) (27), 2.7% (Denmark) (50), 7.4% (Italy) (51), and 4.9% (Michigan, USA) (52).
Yongchen et al. examined the relationship between molecular methods and serological techniques and determined that serological technique was a supplementary method to molecular assay in symptomatic SARS-CoV-2 cases (16). In addition to the above methods, there are antigenic detection methods, which have been already used to diagnose influenza and other respiratory viruses; however, their sensitivity and specificity levels are not accepted. Several studies have been conducted to detect the COVID-19 virus by antigen detection (63, 64). Zhao et al. determined antibody responses to the COVID-19 virus in 137 patients and the sensitivity of diagnostic tests in different days (65), as shown in Table 3. In addition to the above methods, researchers in another study could devise a new method, i.e. a field-based transistor (FET) biosensor, to identify COVID-19. This FET biosensor was designed using graphene sheets with a special antibody against COVID-19 spike protein; hence, this method is a sensitive immunological technique for the detection of SARS-CoV-2 (66). Antigenic methods can be used for the rapid detection of LFA strips with better sensitivity in ELISA. A fluorescent immune-chromatographic LFA technique was introduced in another study to detect the nucleocapsid (N) protein of SARS-CoV-2 (62).
Although ELISA and LFA methods are well-known and used to detect COVID-19viral antigen, there is only one antigen detection test for identify SARS-COV-2, known as the "Sofa 2 SARS antigen test kit" that is approved by FDA (U.S. Food and Drug Administration). This diagnostic kit is used to identify N proteins of both SARS-CoV and SARS-CoV-2 and exploits nasopharyngeal and nasal swab samples.
4.1. Conclusion
The spread of COVID-19 has caused global concern due to its rapid spread in several countries and the progression of this virus's mortality. Therefore, the ability to accurately and fast detect this infectious agent is crucial to saving humans worldwide.
In this article, we tried to examine the diagnostic methods tested by researchers worldwide. According to the findings, although RT-PCR the fundamental diagnosis method of SARS-CoV-2 virus, this technique is a complex process and requires expensive devices. On the other hand, inappropriate sampling leads to false results; therefore, all the methods are useful for the detection of COVID-19 as such, in addition to RT-PCR, other techniques such as serological methods should also be performed.