1. Background
The development of fungal colonies in paranasal sinuses in infected or uninfected sinuses is a common complication (1-6). Chronic sinusitis is one of the most prevalent chronic diseases affecting people of all ages (7). In fact, sinusitis is an inflammation of the paranasal sinuses that may be caused by an infection, allergy, or autoimmune diseases (8). Paranasal sinuses consist of four pairs of air-filled cavities at the entrance of the upper airway, which include maxillary, frontal, sphenoid, and ethmoid sinuses (9). In the beginning, sinusitis is caused by a viral infection that usually lasts more than 10 days, and in 99% of cases, it is completely cured, but in a small number of patients, a secondary bacterial infection may develop, usually caused by aerobic bacteria (10). About 10% of sinusitis cases requiring surgery are caused by a fungal infection. The infection may be caused by a variety of fungal families. Aspergillus fumigatus is the most common cause of fungal infection of the sinuses, and the maxillary sinus is the most common site of such fungal infections. The diagnosis of fungal infections of the paranasal sinuses requires sufficient symptoms (4, 5).
Due to the significance of rapid and accurate diagnosis of fungal infections in patients with chronic sinusitis for the timely treatment and prevention of secondary complications, finding a fast, sensitive, easy, low-cost, and available method seems necessary. Another reason for the need for such a method is the time-consuming nature of fungal culture methods. The classic diagnosis of this Fungusis usually made on the basis of microscopic and culture methods. These techniques are time consuming, relatively inaccurate, and require facilities and experience to interpret the results (6). The usual method of laboratory diagnosis of fungi is to cultivate them, which takes a long time (a few days to several weeks) so that their morphological characteristics are definitely indicative of their species (7). In addition, false-negative results are relatively high in such cases (8). Numerous studies have been conducted on the diagnosis of chronic sinusitis, and different methods have been used for this purpose. Cultivation, immunological, enzymological, and molecular methods have been proposed, each with its own advantages and disadvantages. Non-cultivation methods differ greatly in terms of velocity, reliability, limit of detection, specificity, accuracy, and costs (6).
Few studies have been conducted on the diagnosis of fungal sinus infections using the PCR method (11-17). In 2003, Biswas performed a molecular analysis of the mt cyte b gene sequence in the fungus Cryptococcus neoformans. In that study, he used the primer pair of E1M4 and rE2M4, which were able to detect the fungus. The study showed a high level of diagnosis and specificity (17).
2. Objectives
In the present study, the locus mt cytb with high LOD and specificity was used for the rapid diagnosis of fungal isolates in samples of patients with chronic sinusitis admitted to Rasoul-e-Akram (PBUH) Hospital.
3. Methods
3.1. Samples
Seventy-two samples were collected of maxillary and frontal sinus secretions of patients admitted to Rasool-e Akram (PBUH) Hospital in Tehran for sinus surgery. The samples were collected from patients diagnosed with sinusitis by a hospital Ear, Nose, and Throat specialist while taking into consideration that the disease had reached a chronic stage and the patients had the indications for surgery. Over a nine-month period, 72 specimens were prepared during sinus surgery and poured into vials containing sterile physiological serum.
3.2. DNA Extraction from Fungi
In order to optimize the PCR test and place a positive control for the detection of fungi in clinical specimens, Fusarium solani species was cultivated for 48 hours at a temperature of 37°C in Sabouraud dextrose agar medium, and from the resulting colonies and the 72 samples, DNA was extracted by the boiling method using a DNP kit (Cinnaclon company) based on the protocol.
3.3. Optimizing PCR Test
The primers for the PCR of mt cyt b were E1M4 (Forward: 5´-TGR GGW GCW ACW GTT ATT ACT A-3´) and rE2M4 (Reverse: 5´-GGW ATA GMW SKT AAW AYA GCA TA-3´) according to a former study (14). In this method, to optimize the PCR technique, the suitable concentration and volume of the required components of this method were determined, and finally, the required values were obtained in a PCR test in 25-µL volume. Also, the temperature profile was optimized by the gradient method to mutually amplify the mt cyt b gene as follows: Denaturation at 94°C for one minute, annealing at 50°C for one minute, and an extension at 72°C for two minutes. To obtain better results, we continued the final extension for 10 minutes at the same temperature; this process was performed in 35 cycles. The PCR test was carried out under optimized conditions, and the PCR product was electrophoresed in 1.5% agarose gel containing SYBER Green in 0.5 TBE X5 buffer.
3.4. Cloning PCR Products
After purifying the PCR product, it was cloned using a T/A cloning kit (Fermentas Company) in pTZ57R carrier. The resulting plasmids were extracted by the alkaline lysis method. The PCR method then confirmed the plasmids containing the PCR product (14).
3.5. Limit of Detection
Limit of detection (LOD) test was conducted to obtain serial dilutions from fungal DNA. For this purpose, the extracted DNA of the F. solani fungus was used, and its LOD was measured with a nanodrop device and based on the genome copy number formula and the size of the F. solani genome. In this method, the copy numbers of DNA were known, and serial dilution were prepared.
3.6. Determining PCR Test Specificity
To determine the specificity, we extracted the DNA of seven common microorganisms in sinusitis, such as Staphylococcus aureus, Streptococcus pneumonia, Escherichia coli, Pseudomonas aeruginosa, and viral species, such as hepatitis B virus, herpes simplex virus, and cytomegalovirus, and performed the PCR test for them with a positive and negative control sample.
4. Results
4.1. The Results of Optimization, LOD, and Specificity of PCR
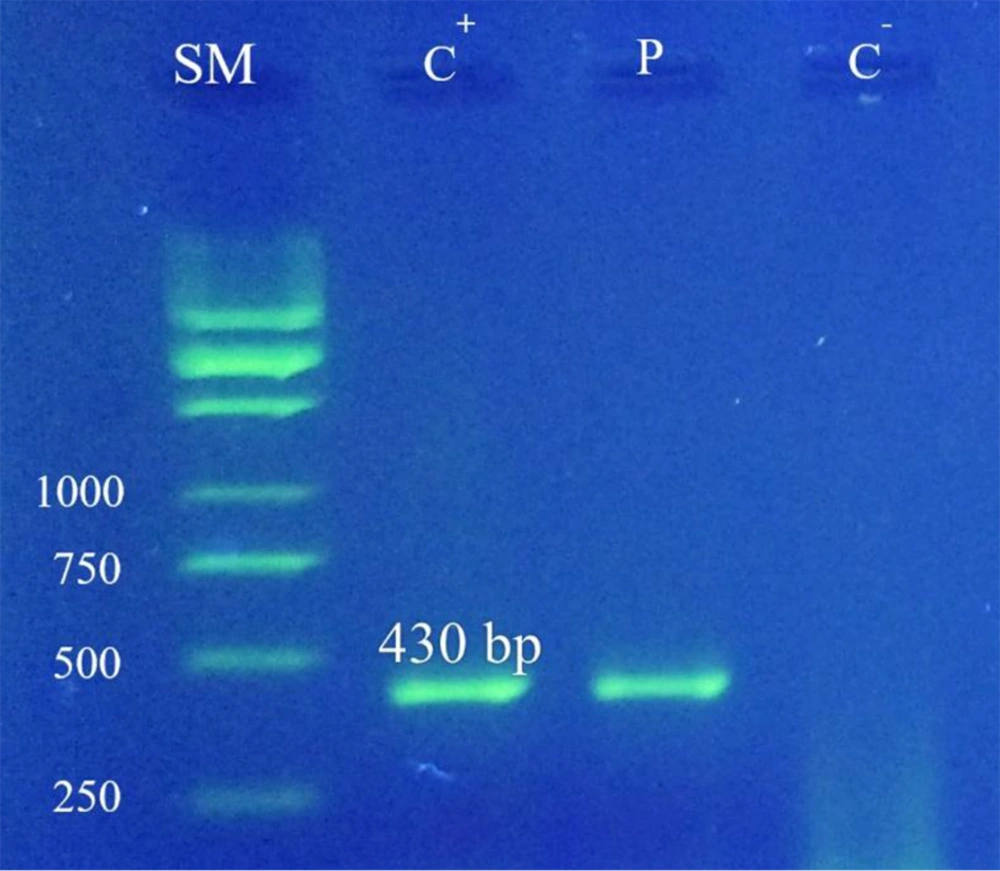
Optimizing the PCR test: After optimizing the PCR test for the concentration of PCR reaction components and thermal profile using universal fungi primers, according to the appropriate thermal program, a PCR cycle was performed on it, and fungal DNA was evaluated along with a negative control sample and a marker size on 1.5% agarose gel. Following the electrophoresis of the PCR product, a band was visualized from the related amplicon with a size of 430 bps (Figure 1). DNA from other fungi, such as Candida albicans, C. neoformans, Aspergillus flavus, Aspergillus parasiticus, and Aspergillus niger, were also used in this experiment, but none of them were amplified with the fE1M4 and rE2M4 primers.
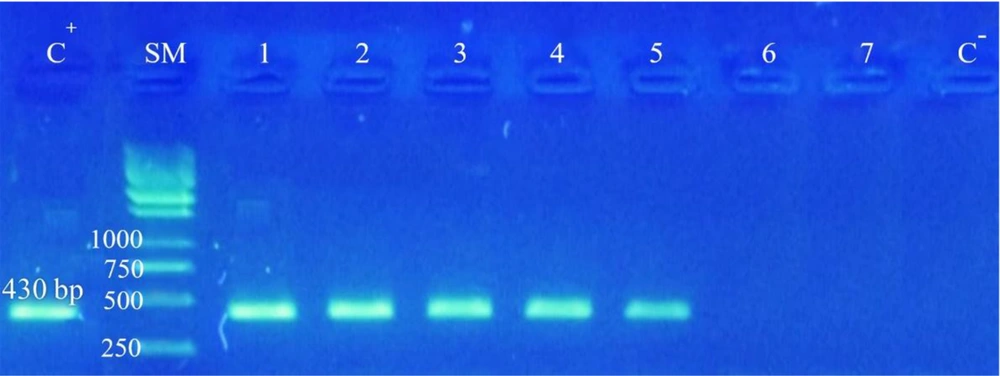
Limit of detection: LOD test was conducted to provide serial dilutions from fungal DNA. Limit of detection findings of PCR test revealed that even if the number of fungal DNA amounts to 50, fungal proliferation occurs. When the number is less than 50, no band could be seen, indicating high LOD of the test (Figure 2).
LOD test. C-: Negative control, C+: Positive control, SM: Size marker kb DNA Ladder 1 (Ferments) 1: 0.1 dilution containing 500 thousand fungal DNA, 2: 0.01 dilution containing 50 thousand fungal DNA, 3: 0.001 dilution containing 5 thousand fungal DNAs, 4: 0.0001 dilution containing 500 fungal DNAs, 5: 10-5 dilution 50 fungal DNAs, 6: 10-6 dilution, 7: 10-7 dilution.
Polymerase chain reaction test specificity: The PCR specificity test was performed using DNA from seven other common organisms in sinusitis, and no unwanted products or bands were observed (Figure 3).
PCR specificity test of Fusarium solani: SM: kb DNA marker size Ladder 1 (Fermentas), C+: Positive control sample (F. solani), 1: Staphylococcus aureus, 2: Streptococcus pneumoniae, 3: Pseudomonas aeruginosa, 4: Escherichia coli, 5: Hepatitis B virus, 6: Herpes simplex virus, 7: Cytomegalovirus, C-: Negative control.
4.2. The Results of Performing PCR Test on Patients’ Samples
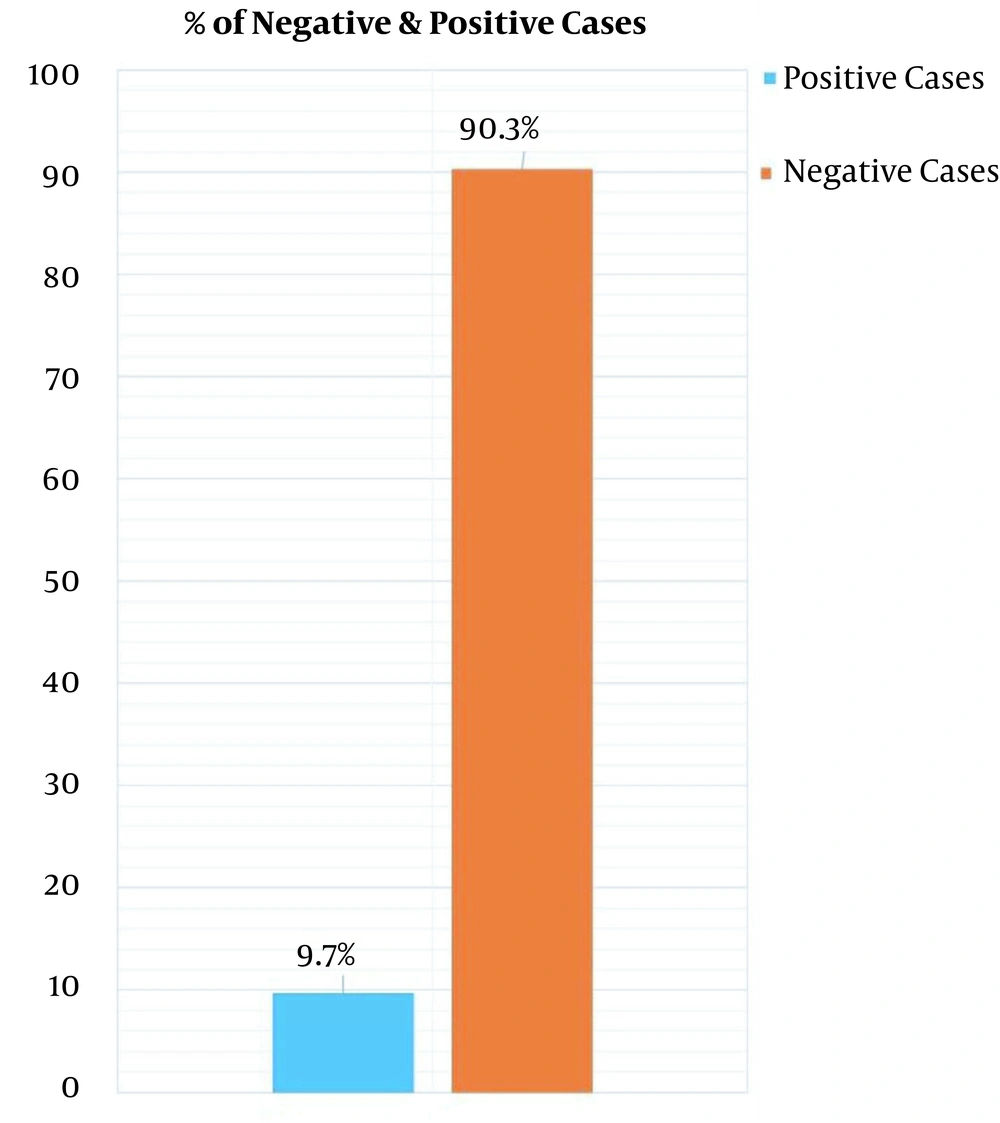
DNA from 72 samples of paranasal sinuses was extracted by the boiling method using a DNP kit (Cinnaclon Company) and was employed for the PCR test. Finally, 7 (9.7%) samples were definitely positive (Figure 4).
Figure 5 shows the results of the PCR test in a number of studied samples.
5. Discussion
Paranasal sinus diseases account for a significant portion of annual cost of treatment (11). Fungal sinusitis is most commonly found in the age groups of 10 to 60 (15). New approaches to the diagnosis of fungal infections and antifungal therapies have emerged based on recent research into the role of fungi in the development and progression of chronic sinusitis. Wang et al., in 1998 designed a primer of the known cytochrome b amino acid sequence to identify and evaluate the racial evolution of Aspergillus pathogens (10). Walter Buzina examined 12 samples of fungi for the molecular detection of Schizophyllum commune. After culturing the samples, they extracted their DNA and the transcriptional sequence of ribosomal DNA s8 / 5 (5.8s rDNA), and ITS region was amplified by the specific fungal primers ITS4 and ITS5, and the products were sequenced by PCR (12). In 2003, Biswas performed a molecular analysis of the mt cyte b gene sequence in the fungus C. neoformans. In that study, he used the primers pair of E1M4 and rE2M4, which were able to detect the fungus. The study had a high level of diagnosis and specificity. In the present study, locus mt cyt b with high a limit of detection and specificity was used for the PCR method. Samples of this study were specifically taken from patients with chronic sinusitis during surgery, and 9.7% of them were diagnosed with fungi (17).
In Iran, Naghibzadeh et al. reported that through the cultivation method, sinus secretions, and light microscopy of 162 patients with chronic sinusitis, 12 patients were confirmed to have fungal infections, which included 2 (1.2%) cases of A. flavus, 9 (5.66%) cases of Alternaria, and 1 (0.6%) case of Paecilomyces. The prevalence of chronic fungal sinusitis among the population community was about 7.4%, which is lower compared to other causes of sinusitis. According to the studies, the most significant pathogen causing the disease in the family of fungi is Alternaria. Compared to other factors causing sinusitis, fungi are less common (16).
Based on the results of the current study, the evaluation of the selected primers with seven various DNA constructs demonstrated 100% specificity (Figure 1). Also, the LOD of optimized test was evaluated for up to 50 fungi (Figure 2). The PCR specificity test results showed that the PCR test had very high specificity and only reacted toward fungal DNA (Figure 3). In addition, out of 72 samples, 9.7% were positive for fungi existence (Figure 4). Samples of this study were specifically taken from patients with chronic sinusitis during surgery, and 9.7% of them were diagnosed with fungi.
As mentioned earlier, fungi are one of the contributing factors of chronic sinusitis. The only way to control the disease is early diagnosis. Therefore, the rapid diagnosis of this microorganism can play an essential role in identifying and controlling the pathogen in early stages of the disease (17).
The PCR technique used in this study to detect different types of fungi was highly accurate, and this method can be used to quickly diagnose the organism in patients with sinusitis and other infections caused by the fungal family (17).
One of the limitations of this study was that the high cost did not allow us to use other diagnostic methods such as culture for comparison at the same time and during molecular detection.
5.1. Conclusions
In the present study, the locus mt cytb was used for the rapid diagnosis of fungal types in samples of patients with chronic sinusitis from Rasoul-e-Akram (PBUH) Hospital. Seventy-two samples were collected from the secretions of maxillary and frontal sinuses of patients during sinus operation. After DNA extraction, the detection of fungi was carried out by amplifying the target sequence using universal primers. At first, PCR was optimized, and LOD and specificity tests were performed. The amplicon was cloned by the T/A cloning method. Based on the results, the evaluation of the selected primers with seven various DNA constructs demonstrated 100% specificity. Also, LOD of the optimized test was evaluated up to 50 fungi. Out of the 72 samples, 9.7% were positive for fungi existence. Samples of this study were specifically taken from patients with chronic sinusitis during surgery, and 9.7% of them were diagnosed with fungi.