1. Background
Recently, Acinetobacte baumannii has become a major threat in the healthcare system due to its high potential to cause nosocomial and opportunistic infections such as catheter-associated urinary tract infections and ventilator-associated pneumoniae. This bacterium is distributed in the environment and is highly prevalent in hospital settings. The bacterium can colonize humans' skin, wounds, and respiratory and gastrointestinal tract and penetrate the lower respiratory tract following aspiration, causing acute pneumonia. Weakness of the immune system, chronic lung diseases, diabetes, long-term hospitalization, and use of urinary catheters and respiratory ventilators are the most important risk factors for increasing the risk of opportunistic infections by this bacterium. This bacterium has different mechanisms for resistance to different types of antibiotics and is usually resistant to most of the available antibiotics.
Reduced membrane permeability, efflux pumps activity, and producing Beta-lactamase and other drug hydrolysis or modifying enzymes are the main resistance mechanisms in A. baumannii, which confer resistance to different antibiotics. There is high plasticity in the genome of this bacterium, which leads to the development of new genes and resistance to disinfectants, heavy metal agents, and antibiotics.
Integrons are mobile genetic elements that carry different antibiotic-resistance genes. Five classes of integrons have been described based on the 5′ conserved segments of the intI gene. In the structure of integrons, the intI gene encodes an integrase, an attI as a recombination site, and promoter gene presence in all types of integrons. Cassettes are promoter-less open-reading frame agents that harbor antibiotic, heavy metal, and antiseptics-resistant genes and can integrate into integrons through site-specific recombination.
Since integrons can transmit antibiotics, heavy metals, and antiseptics resistance genes among bacteria, they can play an important role in the survival and spread of nosocomial pathogens, which in turn increases the cost of treatment and life-threatening nosocomial infections in the hospital setting.
During the past decades, the spread of multidrug resistance (MDR) strain and extensive drug resistance (XDR) strains of A. baumannii have increased, making it a severe public health problem.
The emergence and rapid spread of MDR and XDR strains of A. baumannii are due to the intrinsic resistant genes carried by these strains and also the high capacity of these strains to acquire resistant elements from other bacteria through a horizontal gene transfer mechanism. Accordingly, integrons are critical in spreading resistance genes such as efflux pump genes, beta-lactam resistance genes, and aminoglycoside resistance genes. Based on different reports, class 1 integron followed by class 2 integron are the major contributors to the emergence and spread of the MDR strain of A. baumannii (1-3).
2. Objectives
In this regard, this study was designed to characterize the occurrence of drug resistance and evaluate the prevalence of different integrons as a useful epidemiological tool among A. baumannii isolates taken from patients referred to Ardabil academic hospitals. In the present study, we aimed to determine the antibiotic susceptibility pattern of isolates obtained from academic hospitals, the clonal relationship between clinical isolates, and molecular analysis of isolates with a particular focus on integrons, integron cassette array, and gyrA and parC gene mutations.
3. Methods
3.1. Specimen and Data Collection
In this descriptive, cross-sectional study, a total of 100 A. baumannii isolates were collected from patients admitted to teaching hospitals in Ardabil province during 2017 - 2019. The Institutional Ethics Committee approved this study of the Ardabil University of Medical Sciences (IR. ARUMS.REC.1398.174).
Tryptic soy broth, Nutrient agar, Eosin methylene blue agar (EMB), Sulfide indole motility (SIM), Oxidative fermentative (OF), Simons citrate agar, Triple sugar iron (TSI), and Muller Hinton agar were obtained from Mirmedia company in Iran. All media were prepared according to the instruction of the media container. Primarily, all samples were cultured on EMB agar and incubated overnight at 37˚C. After incubation, all suspected colonies were inoculated into selective and
differential mediums, including TSI, SIM, OF, and Simons citrate agar (4). All isolates biochemically identified as A. baumannii species were subject to molecular confirmation. RecA gene and ITS gene were used for the Acinetobacte genus and A. baumannii species molecular identification (5).
3.2. Antimicrobial Susceptibility Testing
According to CLSI protocols, the disk diffusion method was used to evaluate the antibiotic susceptibility profile of isolates (6). The antibiotic discs were Cefazolin 30 µg, Cefotaxime 30 µg, Ceftazidime 30 µg, Cefepime 30 µg, Imipenem 10 µg, Gentamicin 10 µg, Amikacin 30 µg, Ciprofloxacin 5 µg and polymyxin B (300 µg) (padtanteb, Iran). This study used E. coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 strains as controls for antimicrobial susceptibility testing. Isolates resistant to at least one antibiotic in three or more antibiotic classes were considered MDR strains (7, 8).
3.3. DNA Extraction
The Boling method was used for DNA extraction, and nanodrop was used for calculating the concentration of DNA (9). Polymerase chain reaction (PCR) using highly specific primes was employed for the amplification of genes of interest (Table 1).
| Primers | 3ˊ→ 5ˊ | Product Size (bp) | Annealing°C | References |
|---|---|---|---|---|
| reC-fA1 | CCT GAA TCT TCT GGT AAA AC | 425 | 58 | (5) |
| reC-rA2 | GTT TCT GGG CTG CCA AAC ATT AC | |||
| Ab-ITSF | CAT TAT CAC GGT AAT TAG TG | 208 | 58 | |
| Ab-ITSR | AGA GCA CTG TGC ACT TAA G | |||
| IntI1-F | GGT CAA GGA TCT GGATTTCG | 436 | 60 | (10) |
| IntI1-R | ACA TGC GTG TAA ATC ATC GTC | |||
| IntI2-F | CAC GGA TAT GCG ACA AAA AGG T | 788 | 60 | |
| IntI2-R | GTA GCA AAC GAG TGA CGA AAT G | |||
| IntI3-F | AGT GGG TGG CGA ATG AGT G | 600 | 60 | |
| IntI3-R | TGT TCT TGT ATC GGC AGG TG | |||
| 5′ CS | GGC ATC CAA GCA GCA AG | variable | 59 | |
| 3′ CS | AAG CAG ACT TGA CCT GA | |||
| parC-A-F | ACT GCT TCC GCA TCA ATA C | 919 | 60 | (11) |
| parC-A-R | CAG AAA ACC GCT CTG TAG CG | |||
| gyrA-A-F | AAA CCT GTT CAC CGT CGT TA | 541 | 60 | |
| gyrA-A-R | TAC CGC CTG TAG GGA AGT CA | |||
| aac(6′)Ib-cr-F | TTG CGA TGC TCT ATG AGT GGC TA | 500 | 63 | |
| aac(6′)Ib-cr-R | CTC GAA TGC CTG GCG TGT TT | |||
| qnrA-F | GGA TGC CAG TTT CGA GGA | 500 | 58 | |
| qnrA-R | TGC CAG GCA CAG ATC TTG | |||
| qepA-F | TGG TCT ACG CCA TGG ACC TCA | 1140 | 63 | |
| qepA-R | TGA ATT CGG ACA CCG TCT CCG |
The Sequence of Primers for Amplification of Integrons and Resistance Genes
3.4. Integrons and Gene Cassettes
The existence of integrons and resistance genes was confirmed using the PCR method. The amplification reaction was carried out in a 0.25 µL microtube buffer containing 1.5 µL MgCl2 (25 mM), 0.5 µL dNTPs (100 mM), one µL of each primer (5 pmol/µL, forward and reverse), 0.2 µL Taq DNA polymerase (1 U/µL), double distilled water (DDW = 17/3), and genomic DNA in a final volume of 25 µL. The PCR conditions for amplifying genes of interest were as follows: initial denaturation for 5 min at 94°C, 35 cycles of denaturation at 94°C for 1 min, annealing as presented in Table 3 for every gene for 1min, and extension at 72°C for 1 min. The final extension was carried out at 72°C for 10 min (10). one PCR product of each gene was selected for DNA sequence characterization. The amplicon fragments were separated using electrophoresis on a 1.5% agarose gel with a 100 bp DNA ladder (Parstous, Iran). The agarose gel was visualized under UV following staining with the green viewer (Parstous, Iran). The results of sequences were analyzed with DNAMAN and BLAST software.
3.5. Fluoroquinolone Resistance Genes
Isolates that were resistant to ciprofloxacin disk in the disk-diffusion method were selected. Minimum Inhibitory Concentration (MIC) values (mg/L) for ciprofloxacin were determined using the agar dilution method, and Escherichia coli ATCC 35218 was used as a control standard. Isolates with high MIC were examined for gyrA, parC, aac(6′)Ib-cr, qnrA, and qepA resistance genes. In order to evaluate mutations in gyrA and parC genes, Acinetobacte baumannii (GenBank DQ270238.1, CP049363.1) were used as control sequences, respectively (10, 12).
3.6. Molecular Typing of Isolates Using Enterobacterial Repetitive Intergenic Consensus (ERIC)-PCR
Enterobacterial repetitive intergenic consensus (ERIC)-PCR method was used for molecular typing of Acinetobacte baumannii isolates using a single primer '5′- ATGTAAGCTCCTGGGGATTCAC-3 in a PCR buffer with a final volume of 50 µL containing35.6 µL of sterile deionized water (DDW), five µL of buffer (buffer TBE 10X), three µL (MgCl2), one µL mix dNTP, 0.4 µL Taq DNA polymerase, three µL primer, two µL DNA with the thermal program: one cycle 94°C for 5 minutes, 30 cycles 94°C for 1 Min, 48°C for 1 minute, 72°C for 1 minute and a cycle of 72°C for 10 minutes. The electrophoresis patterns were analyzed and compared using Gelcopmar II v. four software. The Dice coefficient and unweighted pair group method with arithmetic averages (UMGMA) method were used for determining similarity among strains and clustering, respectively (13).
3.7. Statistical Analysis
The chi-square test was used to compare the variables and determine the P-value using SPSS software version 18.0 (SPSS Inc., Chicago, IL, USA). A P-value less than 0.05 was considered statistically significant.
4. Results
4.1. Bacterial Strains
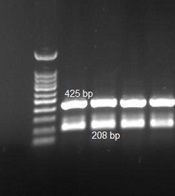
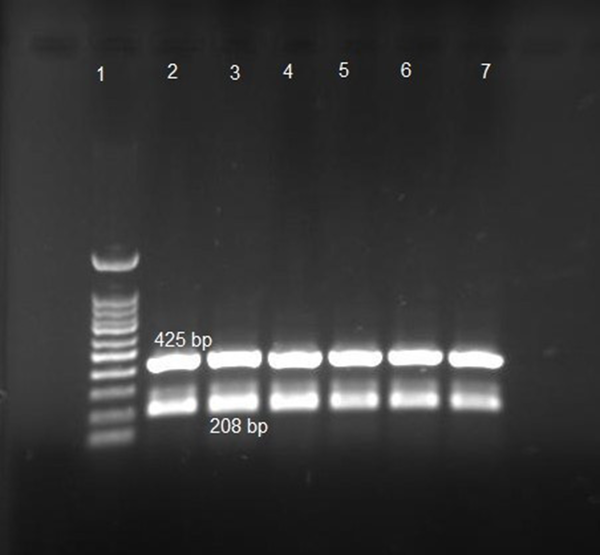
One hundred clinical isolates of A. baumannii from two hospitals in Ardabil province were collected between 2017 and 2019. Acinetobacte genus and A. baumannii species were confirmed by PCR method and genus and species-specific primers. After amplification, the PCR product of the recA gene was sequenced by the Sanger method (pishgam, Iran) and submitted to Genbank under MW092107.1 accession number (Figure 1).
The average age of the patients was 59.81 ± 11.51 61 patients were male (61%), and 39 were female (39%). Based on the length of hospitalization, (n = 8) 8% of the patients were hospitalized for less than one week, (n = 29) 29% of the patients were one week, and (n = 63) 63% of the patients were hospitalized for more than a week.
Hypertension (93%), diabetes (34%), use of urine catheter (23%) and surgery (43%) were the most underlying disease among studied patients. statistical analysis indicated that the mean age of patients (P-value = 0.001) and also the length of hospitalization (P-value < 0.001) were significantly higher in men than women. Most isolates were recovered from ICU (61%) ward, and the trachea was the most prevalent specimen (57%). The frequency of recovered isolates by hospital wards and sources of isolation are presented in Tables 2 and 3.
| Sample Type | No. (%) |
|---|---|
| Urine | 8 (8) |
| Trachea | 57 (57) |
| Lung secretions | 4 (4) |
| Mucus | 1 (1) |
| Blood | 7 (7) |
| Wound | 9 (9) |
| Synovial fluid | 1 (1) |
| Pleural fluid | 6 (6) |
| Catheter | 7 (7) |
Frequency of Isolates Based on Different Samples
| Ward | No. (%) |
|---|---|
| Pediatric | 2 (2) |
| Gynecologic surgery | 5 (5) |
| Urological surgery | 1 (1) |
| General | 17 (17) |
| CCU | 14 (14) |
| ICU | 61 (61) |
Frequency of Samples Recovered from Different Wards of the Hospital
4.2. Antimicrobial Resistance Profile
Antibiotic resistance profile was evaluated with disk diffusion with a panel of 10 antibiotics according to CLSI guidelines (6), and E. coli atcc 25922 was used as quality control. In this study, Acinetobacte baumannii strains showed high resistance to all antibiotics except polymyxin B, with the highest antibiotic resistance in cefazolin (100%), ciprofloxacin (99%), imipenem (99%), cefotaxime (98%), meropenem (98%), Ceftazidime (97%), and to a lesser extent cefpime (89%), gentamicin (82%), and amikacin (78%). All strains in this study were sensitive to polymyxin B (100%) (Table 4).
| Susceptible | Intermediate | Resistant | |
|---|---|---|---|
| Cefazolin | 0 (0) | 0 (0) | 100 (100) |
| Ciprofloxacin | 1 (1) | 0 (0) | 99 (99) |
| Imipenem | 0 (0) | 1 (1) | 99 (99) |
| Cefotaxime | 0 (0) | 2 (2) | 98 (98) |
| Meropenem | 1 (1) | 1 (1) | 98 (98) |
| Ceftazidime | 2 (2) | 1 (1) | 97 (97) |
| Cefepime | 6 (6) | 5 (5) | 89 (89) |
| Gentamicin | 9 (9) | 9 (9) | 82 (82) |
| Amikacin | 13 (13) | 9 (9) | 78 (78) |
| Polymyxin B | 100 (100) | 0 (0) | 0 (0) |
The Results of the Antibiotic Susceptibility Pattern of the Studied Isolates Using the Disk Diffusion Method a
4.3. Integron Assessment of Acinetobacte baumannii Isolates
Integron and cassette carriage were detected using the PCR method, and amplicons of integron class 1 and cassette one were sequenced by the Sanger method (genomin, Iran). Sequences were edited and analyzed using DNAMAN and BLAST software, and the consensus sequences were submitted to Genbank. (accession no. MT891123, MT891124). Class 1 and 2 integrons were verified in 70 (70%) and 21 (21%) isolates, but class 3 integron was not found in any of the strains. The presence of an 1100 bp cassette in one segment with the aadA1 gene was detected in all integron1 carriage strains (100%).
4.4. Ciprofloxacin Susceptibility and Amino Acid Substitutions
Isolates resistant to ciprofloxacin disk were selected, and ciprofloxacin MIC was measured for them. The MIC were ranging from 2 (µg/mL) (1%), 32 (1%), 128 (11%), 256 (45%), 512 (35%), to ≥ 1024 (µg/mL) (6%). Strains with high MIC (≥ 1024) were evaluated for mutations in DNA gyrase subunits, topoisomerase, and the presence of resistance genes, including aac(6′)Ib-cr, qnrA, qepA. Amplicons of gyrA and parC were sequenced, and such as integron and cassette were analyzed. The gene parCgene has not any mutation, while in the gerA gene, a single substitution (S83L) was detected in the QRDR region (accession no. 2494853, 2494855 under review in Genbank). Also, Ib-cr aac (6%) gene was observed in 2 strains, but qnrA and qepA genes were not found in either strain (accession no. 2478338).
4.5. Molecular Typing of Acinetobacte baumannii Isolates
Molecular typing of about 100 isolates of A. baumannii by ERIC-PCR has been presented in Figure 1. In the analysis of the ERIC-PCR profile, 24 clusters were identified that the band size was between 300 and 1000 bp, and the number of bands was between 3 - 9. As can be seen in the dendrogram, there are two large clusters
Both clusters belong to Imam Khomeini Hospital in Ardabil, and the patients were admitted to the ICU ward. In Cl-A, isolates were recovered from patients hospitalized in the ICU for more than one week, while the members of Cl-B were isolated from patients hospitalized for up to one week.
Cl-A members were isolated from the trachea, urine, and wound samples, while Cl-B members were isolated from the trachea, blood, wound, sputum, and urine samples. The age of patients in both clusters was high (< 50 years), and no significant difference was observed in terms of the age of patients in both clusters (P > 0.05). Gender differences in both clusters were significant, and most patients involved in this cluster were women.
5. Discussion
Acinetobacte is a non-fermenting gram-negative bacillus that has so far identified more than 30 species in the genus Acinetobacte. Most of these species are environmental bacteria and are not associated with human infections. A. baumannii, A. calcoaceticus, and A. lwoffii are the most common species associated with human infections. Also, A. junii has recently been isolated
as an opportunistic pathogen in cancer patients (14).
Outer membrane protein A (OmpA) (making biofilm), polysaccharide capsule (anti-complement activation agent), and fimbriae (attachment agent) are the most important virulence factor which confers high resistance to the harsh environment (15).
Acinetobacte species, especially A. baumannii, are important pathogens of nosocomial infections with high mortality. A. baumannii is a major health problem, especially in the ICU and NICU wards, and can infect these wards for a long time and involve immunocompromised patients (16).
In the present study, 100 non –repetitive A. baumannii isolates were collected from a teaching hospital in Ardabil province. Isolates were recovered from patients admitted to ICU sector and also, hypertension, duration of hospitalization, and gender of patients were the most important predisposing factors to A. baumannii infections, which was consistence with another study in Iran and china (17, 18).
A. baumannii uses different mechanisms to develop resistance to different classes of antibiotics and is approximately resistant to a wide range of antibiotics. Over the past decades, MDR strains have increased, and treatment options for treating infections caused by the bacterium have increasingly dropped. Three main mechanisms have been identified that mediated resistance to quinolones: (1) mutation in antibiotic target (2) efflux pump (3) Qnr protein plasmid.
The most common known mechanism for resistance to quinolones in gram-negative bacilli is mutations in the target enzyme, including DNA gyrase and topoisomerase IV, encoded by the gyrA and gyrB, parC, and parE genes, respectively. In A. baumannii, rapid resistance to ciprofloxacin and nalidixic acid associated with chromosomal mutations in the Quinolone resistance determining regions (QRDRs) are from the gyrA and parC genes (11).
Changes in the efflux pump lead to resistance to a wide range of antibiotics, including tetracycline, chloramphenicol, fluoroquinolones, and trimethoprim. Quinolones are bacteriostatic antibiotics targeting the enzyme DNA gyrase, inhibiting DNA replication and transcription (19).
The aim of the present study was to investigate the prevalence of the MDR strain of A. baumannii and determine of involved mechanisms of resistance to fluoroquinolone in clinical isolates of A. baumannii taken from patients admitted to teaching hospitals in the Ardabil province of the northwest of Iran. Therefore we assessed the presence of mutations in the gyrA and parC genes and the presence of plasmid-resistance genes, including the Ib-cr aac gene, qnrA, and qepA.
A single substitution (S83L) in the gyrA gene and
the presence of Ib-cr aac were detected in isolates with high MIC to ciprofloxacin (≤ 1024). Single amino acid substitution in GyrA (Ser83Leu) and ParC (Ser80Leu) are associated with high-level resistance to ciprofloxacin and nalidixic acid
Two isolates that were recovered from the trachea and catheter had high MIC to ciprofloxacin (≤ 1024)
which were screened for gyrA, parC, and PMQR genes by PCR and sequencing in which mutations in the gyrA gene and the presence of aac(6′)-Ib-cr resistance gene were detected, respectively. qnrA and qepA genes were not found in the two isolates.
Qnr A, aac(6′)-Ib-cr and qepA are plasmid-mediated fluoroquinolone resistance genes, and the presence of these genes increases the MICs of ciprofloxacin.
qnrA encodes a protein that protects topoisomerase II from quinolones, and AAC (6′)-Ib-cr gene encodes an aminoglycoside acetyltransferase which modifies quinolone drugs. qepA gene is related to the expression of the active efflux pump (20). Plasmid-mediated quinolone resistance (PMQR) is the group of genes associated with decreasing susceptibility to quinolones such as norfloxacin and ciprofloxacin (12).
In this study, the frequency of MDR isolates was 99%, and the study of 3 classes of integrons showed that the frequency of class 1 integrons is high, which in turn plays an important role in the greater and easier spread of antibiotic resistance. Cassette 1 sequencing showed the presence of the aadA1 gene encodes
aminoglycoside 3''-O-nucleotidyltransferase, which confers resistance to the aminoglycoside antibiotics such as streptomycin and spectinomycin (21). The results of this study indicated that a single substitution (S83L) in gyrA gene and Ib-cr aac play a major role in conferring a high level of resistance to ciprofloxacin.
In our study, the resistance ratio to each tested antimicrobial agent was over 90%, and only polymixin B was able to eliminate resistant isolates. Despite previous studies, in this study, the resistance rate to imipenem and meropenem was very high (22).
Integrons can carry different cassettes that confer resistance to several classes of antibiotics. In this study, 70% of the isolates had class 1 integrons, 21% had class 2 integrons, and no class 3 integrons were identified. Casstte array of integron 1 showed aadAgene. In another study in northwest Iran, the prevalence rate of class 1 integron in multidrug-resistant Acinetobacte baumannii (MDRAB) clinical isolates was 92.5% (23).
In a study conducted by Goodarzi et al. in Tehran, the resistance rate among isolates was between 39.3% and 99.1%, and similar to our study, no resistance to polymyxin B was observed (24). In this study, the resistance rate to imipenem and meropenem was 99.1% and 62.5%, unlike another similar study in Tehran, the resistance rate for imipenem was 4.5% (25). Consistency with Goodarzi's study: In our study, the resistance rate to carbapenems was more than 98%, which is remarkable.
During the past decades, the prevalence of multidrug and extensively drug resistant A. baumannii strain has become a serious threat in hospital setting. So recently, this bacterium has been one of the most important causes of ventilator-associated pneumonia and sepsis in patients in intensive care and neonatal intensive care units. The bacterium's resistance to harsh environmental conditions, antibiotics, and disinfectants and its association with fatal nosocomial infections have made it a serious threat. Integrons are mobile genetic elements that confer resistance to the different classes of antibiotics through a horizontal gene transfer pathway. Structurally, they can obtain different cassettes which encode different antibiotic-destroying enzymes and have a critical role in antibiotic resistance distribution among different species of bacteria. Recently, the relationship between multiple drug resistance, MDR strain, and integrons is well documented.
In the present study, the trachea was the main A. baumannii isolation source (56%) which was in accordance with previous studies in which respiratory specimens are prominent (26).
Although Carbapenems such as imipenem and meropenem are the drug of choice for the treatment of serious nosocomial infections caused by Acinetobacte, carbapenem-resistant strains are increasingly encountered. However, the distribution of carbapenem-resistant strains of AB differs over the different geographical regions. At present, the resistance rate to carbapenems is higher than the other country like Turkey (80%) (27), Germany 3.5% (28), and Russia (45%) (29). In critical conditions, combination therapy such as meropenem/colistin has been recommended (30).
This study showed that in Ardabil province, such as in other parts of Iran, there is high antibiotic resistance among A. baumannii strains, and all strains are MDR.also, more than half of the strains and one-third of them harboring integron class 1 and 2, respectively. The most common site of A. baumannii isolation was the ICU sector which requires more attention to this ward
and the patients hospitalized in this site.
5.1. Conclusions
In the past few years, carbapenems have been the most effective drug against antibiotics, but the results of this study show that the resistance rate for carbapenemase in teaching hospitals in Ardabil has reached an alarming state. As a result of prolonged exposure to these valuable drugs and the use of inappropriate doses, resistance has developed initially. Therefore appropriate use of effective antibiotics, multiple drug combinations, and isolation of infected patients with drug-resistant strains in hospitals from the rest of the patients may help control the emergence of resistance.