1. Background
Papillomaviruses sometimes cause infections of the skin and mucosal membrane, which leads to the emergence of different types of warts (cutaneous, plantar, flat, anal, genital, and laryngeal) or many cancers (uterus, penis, and vulva cancers) (1). The leading cause of human papillomavirus transmission is sexual contact. Various types of this virus are preferentially associated with some clinical lesions. One of the most common types of cancer worldwide is cervical cancer (1). Cervical cancer (CC) has the fourth highest incidence and fatality rate of any malignancy globally. On the other hand, it is the leading cause of female cancer and death in many low-resource nations. Cervical cancer develops slowly, taking years to decades (2). The finding that human papillomavirus (HPV) infection is intangible with CC development was a watershed moment in preventing this disease from causing CC development.
E6 and E7 are the two major carcinogenic proteins produced by the HPV cycle (3). These genes' products bind to and inactivate tumor suppressor genes, causing the host cell cycle to be disrupted. Twelve HPV genotypes have been designated as high risk (HR-HPV): 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, and 59. More than 60 genetic types of HPV have been described. There are strong associations between certain genotypes and clinical lesions (4, 5). Human papillomavirus is the most studied and well-known. Human papillomavirus has around 150 different types (6). Real-time PCR can distinguish nearly related sequences by combining simultaneous PCR amplification with extensive computer analysis of the collected kinetics data (7). Fluorescent dyes are used to detect the amplified product in real-time PCR. These dyes are linked to PCR primers and oligonucleotide probes, which selectively adhere to the amplified product during thermocycling, allowing for accurate detection of viral presence in the sample (8). As a result, real-time PCR has been used in screening programs because it provides a highly sensitive method for qualitative identification and genotyping of HR-HPV types in clinical samples (9). The cellular biology of HPV supports a primary role in the development of neoplasia (10). The viral types most closely associated with CC, particularly HPV-16 and 18, can transform cells in the culture to lose their normal growth control mechanisms. The DNA from these types integrates into host DNA (10, 11).
2. Objectives
The goal of this study was to find HPV 14, 16, 18, and 45 genotypes in urogenital swabs by using a real-time PCR amplification test for quantitative genotyping of HPV DNA types 16, 18, and 45 and for simultaneous quantitative detection of HPV DNA types 31, 33, 35, 39, 51, 52, 56, 58, 59, 66, and 68, for a total of 14 HPV genotypes.
3. Methods
Inclusion Criteria: Samples were used from women with CC confirmed by some tests recommended by the doctor.
Exclusion Criteria: Contaminated samples were excluded, as were individuals who received treatment before taking the sample.
This is a case-control study. Eighty-six cervical swabs from Iraqi women referred by the Al-Yarmook teaching hospital in Baghdad, Iraq, were included in the study. The ages of cases varied from 23 to 70 years, and specimens were obtained between March 2020 and March 2021. A doctor completed a case investigation form with information specific to each patient, including personal data, gynecological history, Pap smear, and histological examination results.
3.1. Sample Collection
Cervical swabs were taken per guidelines. For sampling, a cervical brush (1 - 1.5 cm) was inserted into the cervical canal after removing any excess mucus from the area around the ectocervix. The brush was then rotated three full turns in a counterclockwise direction before being removed from the canal and inserted into a 2 mL nuclease-free tube with 0.5 mL of the transport medium (Sacace), leaving the brush end inside the tube. Swab samples were frozen at -20 - 80°C or kept at 2 - 8°C for no more than 48 hours.
3.2. DNA Isolation
DNA was extracted from the vaginal swabs using the commercial kit DNA-Sorb-A (Sacace, REF K-1-1/A) following the manufacturer's instructions. After the DNA was successfully extracted, the eluted DNA was stored at -20°C (12).
3.3. Detection and Quantification of HPV
Detection protocol was done using the commercial Kit V31-100/F FRT and as follows: For each sample reaction (including clinical samples and controls), 10х(N+1) μL of PCR-mix-1-FRT HPV 14 was added into a new tube. Then, 5.0х(N+1) μL of PCR-mix-2 buffer and 0.5х(N+1) μL of TaqF DNA polymerase were added. The tubes were then vortexed. Finally, 10 μL of DNA sample was added from test or control samples to the prepared tubes (13).
3.4. Statistical Analysis
Statistical analysis was performed using SPSS and Excel 2016 software.
4. Results
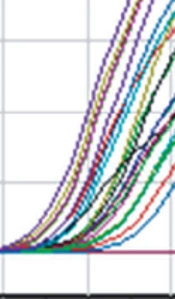
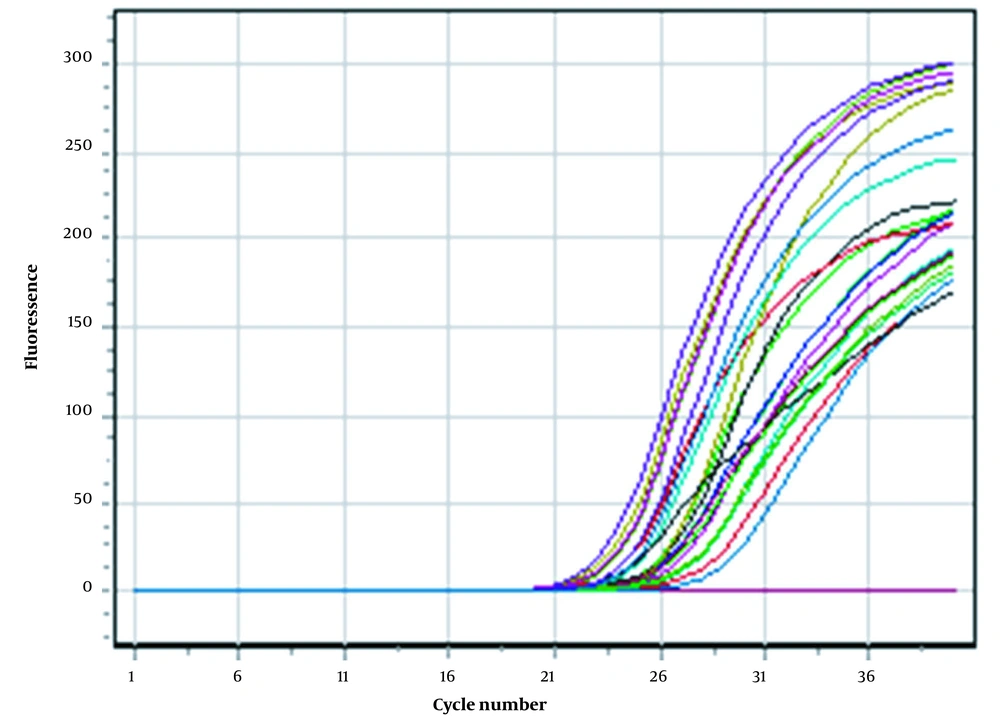
After the completion of the real-time PCR, the amplification results for all samples are shown in Figure 1. Each curve represents the amplification of the desired target region of the virus. The test was declared successful if the negative controls showed no positive fluorescence signal and the standards (Fam, Joe/Hex, Rox, Cy5, Cy5.5) showed positive signals in all channels.
In this study, 14 HPV genotypes were assayed, and the resulting frequencies are shown in Table 1. Genotype 16 showed the highest frequency, followed by genotypes 45 (22%), 18 (14%), 35 (6%), 59 (6%), 52 (4%), and 58 (4%). Genotype 31 showed a 2% frequency, while genotypes 33, 39, 51, 56, 66, and 68 showed the lowest frequency (1%).
| Genotype | Number of Cases | Frequency |
|---|---|---|
| 16 | 25 | 29.06977 |
| 18 | 14 | 16.27907 |
| 45 | 19 | 22.09302 |
| 31 | 2 | 2.325581 |
| 33 | 1 | 1.162791 |
| 35 | 6 | 6.976744 |
| 39 | 1 | 1.162791 |
| 51 | 1 | 1.162791 |
| 52 | 4 | 4.651163 |
| 56 | 1 | 1.162791 |
| 58 | 4 | 4.651163 |
| 59 | 6 | 6.976744 |
| 66 | 1 | 1.162791 |
| 68 | 1 | 1.162791 |
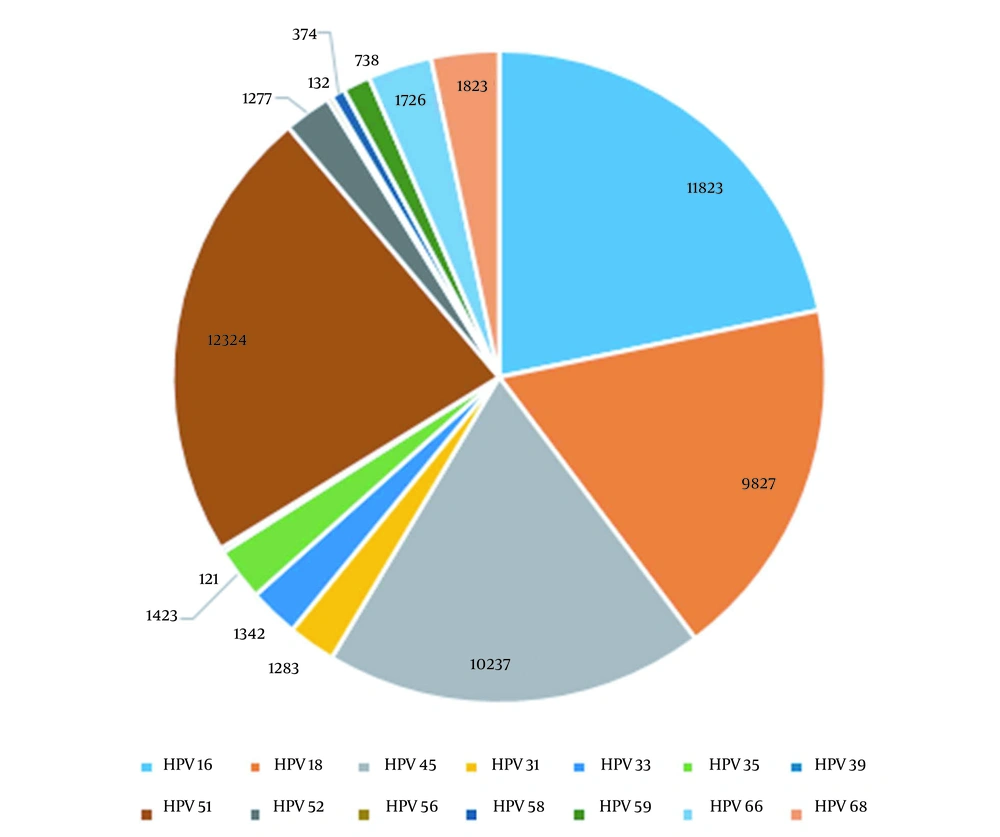
The sensitivity of the PCR-Tm analysis assay for each genotype and their corresponding mean copy numbers are seen in Table 2 and Figure 2.
| Genotype (HPV) | Copy Numbers |
|---|---|
| 16 | 11823 |
| 18 | 9827 |
| 45 | 10237 |
| 31 | 1283 |
| 33 | 1342 |
| 35 | 1423 |
| 39 | 121 |
| 51 | 12324 |
| 52 | 1277 |
| 56 | 132 |
| 58 | 374 |
| 59 | 738 |
| 66 | 1726 |
| 68 | 1823 |
5. Discussion
An estimated 90% of all HPV infections go away on their own within two years after they are contracted. In a small percentage of cases, certain HPV infections can persist and progress to cancer (14)
In research conducted by Alizi et al. (15), the prevalence of HPV among Iraqi women was very low (9%). These findings were consistent with an earlier study in Iraq by Faik et al., which showed 12.38% (16). Comparing Iraqi populations to those in industrialized countries, their Islamic nature and moral dedication to their country may explain the low incidence of HPV and other sexually transmitted illnesses. It has been determined via several examinations in Iraq that virus type 16 is the most prevalent variant in the country (17).
Other investigations used a Real Time amplification test for qualitative detection and genotyping of human papillomavirus (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68) in urogenital swabs and biopsies (Sacace, Italy), and related methodologies yielded comparable results (18, 19).
A study from Iraq found that HPV 33 (18%) was the most common type, and the other found HPV 16 (58.3%) as the most common type, followed by HPV31/33 (25%) and HPV18 (16.7%), which are consistent with our findings (16, 20). The disparity in HPV genotype prevalence may be attributable to sample size, geographic differences, utilizing various techniques, and different primer pairs.
According to He and He (21), the overall rate of HR-HPV infection was 12.6% in 17,319 women, and the five most prevalent types were genotypes 52, 53, 58, 16, and 56, which agrees with our findings.
Similar frequencies were observed in Tianjin (12.5%) (22), Guangdong (7.3%) (23), and Beijing (7.0%) (24) in China. The most frequent HR-HPV types in Sichuan (52, 53, 58, 16, 56) did not appear to be the same as in other Chinese cities (13). In other countries, studies have found that HPV frequency is 12.2% in Canada (25), 34.5% in Peru, 10.3% in Iran, and 9.3% in Australia (26-28).
Aoyama-Kikawa et al. found that HPV infection was present in just 4.6% of Japanese women (29). These results confirm Becker et al. that HPV prevalence varies by geography and ethnicity (30). The only exception is HPV 66, classified as "possibly" carcinogenic (31-33). HPVs 16 and 18 are the most frequent high-risk genotypes worldwide, accounting for 75% of all cervical cancer cases and HPV45 (34, 35).
5.1. Conclusions
The real-time PCR method efficiently detected and genotyped HPV-DNA, which could help in earlier detection and clinical care of HPV-infected patients by reducing costs and workload.