1. Background
Mycobacterium simiae, a slow-growing photochromogenic non-tuberculosis mycobacteria (NTM), is considered the most prevalent and well-known mycobacteria among the M. simiae complex. Mycobacterium simiae was first detected in a Macacus rhesus monkey in 1965, and the disease was identified in humans several years later (1-4).
As the only niacin-positive NTM similar to Mycobacterium tuberculosis, and with a strong potential of causing pulmonary diseases in both healthy and human immunodeficiency virus (HIV)-positive patients, early differentiation between M. simiae and M. tuberculosis is an essential step in effective M. simiae therapy (5-8). Epidemiologically, among NTM species, M. simiae ranks as the most frequent clinical slow-growing mycobacterium in Iran, while among other Middle Eastern countries, M. simiae ranks the second (9-11).
Despite the clinical and epidemiological importance of M. simiae worldwide, including in Iran, there is no clear and effective treatment regimen for M. simiae and its different subtypes (12, 13).
2. Objectives
3. Methods
3.1. Setting and Samples
In this study, all sputum samples with M. simiae confirmation submitted to the National Reference TB Laboratory of Iran from May 2014 to May 2016 were included. All the included clinical isolates from patients with NTM met the American Thoracic Society (ATS) and Infectious Diseases Society of America (IDSA) diagnostic criteria and were considered as the causative pathogen for the disease. Based on the Petroff method, digestion and decontamination of all sputum samples were performed using 4% NaOH and 1 N HCL (16). Sputum smear and culture were performed for all patients. For culture, the sediments were inoculated onto three Lowenstein-Jensen (L.J) mediums and were incubated for 12 weeks. The inoculated cultures were observed twice per week to control and confirm M. simiae (1, 9).
The study was approved by the National Research Institute of Tuberculosis and Lung Diseases (NRITLD), Shahid Beheshti University of Medical Sciences, Tehran, Iran (ethics code: IR, SBMU, NRITLD, REC, 1396, 355).
3.2. DNA Extraction
Mycobacterial DNA was extracted by a commercial Qiagen DNA extraction kit (QIAamp DNA Mini Kit; Cat. No 51306, Hilden, Germany) (17-19). The concentration of the extracted DNA was measured by Pico 100 Spectrophotometer (Saffron Walden, UK; Version 4.0/21/03/11). Then, samples were stored at -20°C and were transported to the molecular department using cold boxes.
3.3. Molecular Genotyping of Isolates
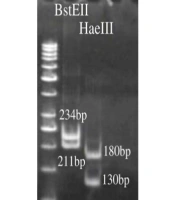
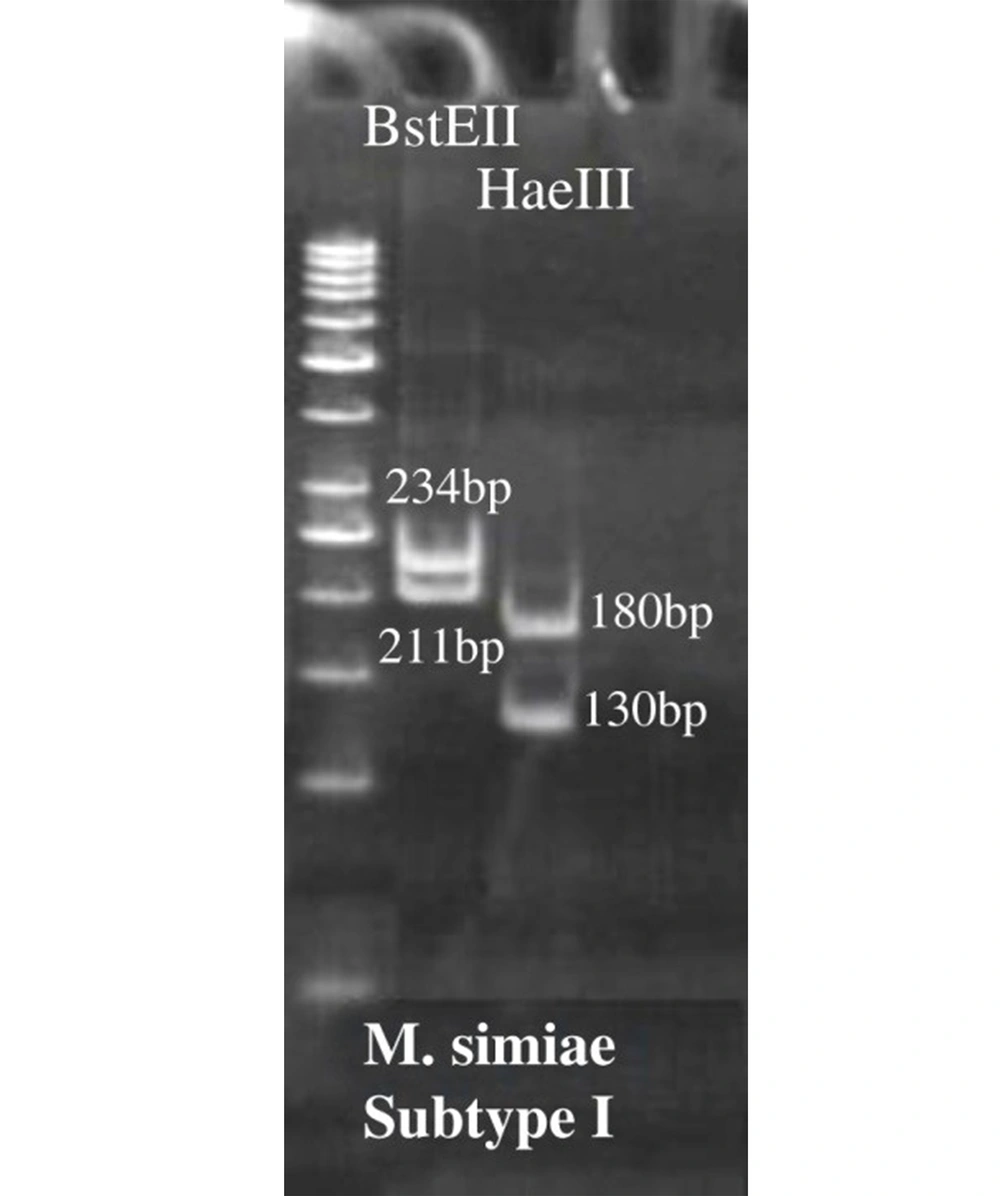
As previously described, hsp65 gene spacer polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) assay was selected for rapid and accurate identification of M. simiae patients (20). In this regard, a set of primers, including TB15 and TB17, followed by TB11 and TB12 were used to amplify a 439bp fragment after two consecutive PCRs. Primer details and targeted regions are summarized in Table 1. Polymerase chain reaction for the first step was amplified in a 25 μL PCR mixture containing 4 pmol of primers TB15 and TB17, 1 μL deoxynucleotide triphosphates, 1.5 μL MgCl2, 0.25 U Taq DNA polymerase, 2.5 μL dimethyl sulfoxide (DMSO), 5 μL 10X PCR buffer and 5 μL (20 ng) of extracted DNA. For the second step, 5 μL (20 ng) of PCR product of the previous step was added to a 50 μL PCR mixture consisting of 8 pmol of primers TB11 and TB12, 1 μL deoxynucleotide triphosphates, 1.5 μL MgCl2, 0.25U Taq DNA polymerase, 2 μL DMSO, and 5 μL 10X PCR buffer. The amplified products were digested by HaeIII and BstEII restriction enzymes and then incubated overnight at 37°C. With slight modification, digested fragments were run on an 8% polyacrylamide gel electrophoresis (21).
| Allele Specific Primer (5’ - 3’) | Paired Primer (3’ - 5’) | Detection Target | Length of Polymerase Chain Reaction Product (bp) | ||
|---|---|---|---|---|---|
| TB15 | CGT AYG ACG AAG AGG CCC GT | TB17 | WAS GGR TCC TCS AGG ACS GC | Hsp65 gene | 470 |
| TB11 | ACC AAC GAT GGT GTG TCC AT | TB12 | CTT GTC GAA CCG CAT ACC CT | Hsp65 gene | 439 |
| rpoB516 | CAGCTGAGCCAATTCATGGA | RIRm | TTGACCCGCGCGTACAC | rpoB516 | 218 |
| rpoB526 | CTGTCGGGGTTGACCCA | rpoB526 | 185 | ||
| rpoB531 | CACAAGCGCCGACTGTC | rpoB531 | 170 | ||
| katG315 | ATACGACCTCGATGCCGC | katGOF | GCAGATGGGGCTGATCTACG | katG315 | 292 |
| inhAP-15 | GCGCGGTCAGTTCCACA | inhAPF2 | CACCCCGACAACCTATCG | mabA-inhA:-15 | 270 |
| RRS1539 | GGGGCGTTTTGCTGGTGCTCC | RRS1096 | GCGCAACCCTTGTCTCATGTTG | rrs gene | 300 |
| GYRA f | CAGCTACATCGACTATGCGA | GYRA r | ATGAGGTACACCGAAGCCC | gyrA gene | 320 |
Details of Targeted Regions, Length of Polymerase Chain Reaction Products, and Primers Used in the Study
Identification of M. simiae infected cases was performed using algorithms proposed by Roth et al. and Telenti et al. (21, 22). Then, the digested fragments were compared with those of patterns deposited in a freely available database (http://app.chuv.ch/parasite/index/html) for subtype classification of M. simiae isolates.
3.4. Molecular Drug Susceptibility Testing
Using the national surveillance data of Iran, we had access to limited medications in Iran due to sanctions. Considering the ATS recommendation to find the best accessible, effective, and easy-to-use drug regimen, the most common and frequent anti-mycobacterial antibiotics in Iran, including rifampin (RIF), isoniazid (INH), amikacin (AMK), kanamycin (KAN), and ciprofloxacin (CIP) were selected for in vitro investigations. The specific mutant codons and genotyping of drug-resistant patterns have been described in previous studies (23-25). So, mutations at rpoB codons 516, 526, and 531 confirmed resistance to RIF. Also, any mutations at katG codon 315 and inhA-mabA promoter region were considered INH resistance. For both AMK and KAN, mutations of the rrs gene (at positions 1400 and rarely 1401) resulted in the presence of M. simiae resistant cases, while gyrA mutations at positions 90, 91, and 94 demonstrated resistance to CIP.
We evaluated the susceptibility of M. simiae subtypes to RIF and INH through performing a multiplex-PCR targeting rpoB and both inhA and katG genes, respectively. Polymerase chain reactions for both antibiotics were amplified in a 25 μL PCR mixture containing 10 pmol of each specific primer (Table 1), 1 μL deoxynucleotide triphosphates, 4 μL MgCl2, 1.25 U Hot Start Taq enzyme, 0.5 μL DMSO, 2.5 μL PCR Buffer, 0.25 μL UNG, and 5 μL (20 ng) of extracted DNA. Amplification cycles were divided into three steps: Initial denaturation at 95°C for 5 min, 40 annealing cycles at 68°C and 70°C for RIF and INH, respectively, for 30 sec, followed by a final extension at 72°C for 5 min. Finally, 7 μL of PCR products were examined for banding patterns by 8% polyacrylamide gel electrophoresis.
In RIF susceptible cases, the 218-bp fragment was represented by rpoB codon 516-specific PCR product, the 185-bp fragment was represented by rpoB codon 526-specific PCR product, and the 170-bp fragment was represented by rpoB codon 531-specific PCR product. Susceptibility to RIF was confirmed in the group of M. simiae patients who represented all specific fragments. In addition, in case of a mutation existing in a given codon or region, no related allele-specific PCR was generated, which illustrated resistance to RIF.
For INH, the 270-bp band was represented by the katG codon 315-specific PCR product, whereas the 292-bp fragment was represented by the inhA-mabA promoter region of the specific PCR product. Mutations in any region confirmed INH resistance, while the susceptible patients were represented by 292 and 270 bp fragments (24).
Susceptibility to AMK/KAN was examined using 10 pmol of a set of primers RRS1539 and RRS1096 (Table 1). To amplify the 300-bp fragment of the rrs gene, 5 μL (20 ng) of extracted DNA was added to a PCR mixture containing 1 μL deoxynucleotide triphosphates, 10 μL MgCl2, 1.25 U Taq enzyme, 0.5 μL DMSO, 5 μL PCR Buffer, and 0.25 μL UNG (Total volume: 50 μL). An initial denaturation accomplished the amplification at 95°C for 7 min, 40 cycles at a specific annealing temperature of 60°C for 60 sec, and DNA extension at 72°C for 5 min. To identify the most frequent mutations, the 300bp PCR product was digested with endonucleases DdeI and TailI for 16 h at 37°C and then was visualized by ethidium bromide staining and UV light. Enzymatic digestion displayed 248 and 191 bp fragments using DdeІ endonuclease. Moreover, digestion with TailІ enzyme was represented by 187 and 154 bp bands. In mutant cases, the lack of 248 or 154 bp fragments was apparent and indicated AMK and KAN resistance fields (23).
Susceptibility to CIP was examined by a single PCR that targets the gyrA gene and performs single strand confirmation polymorphism (SSCP) gel electrophoresis. A 50 μL of PCR mixture, including 2 μL deoxynucleotide triphosphates, 1.5 μL MgCl2, 1.25 U Taq enzyme, 2 μL DMSO, 2.5 μL PCR Buffer, and 10 pmol of GYR-A forward and reverse primers (Table 1) was amplified to present a 320bp fragment. Amplification was completed by initial denaturation at 95°C for 5 min, eight consecutive annealing cycles at five steps (the first step was started at 95°C, 60°C, and 70°C, and for the entire cycles, just the middle 60°C was reduced a degree step by step) for 1 min. Also, a final extension of fragments was performed at 72°C for 7 min. After 10 min more of DNA denaturation at 95°C, the amplified fragments were visualized using silver staining. GyrA mutations at positions 90, 91, and 94 were detected by loading a wild-type H37Rv on the same SSCP to compare patterns of M. simiae strains in contrast with the wild-type patterns at the gyrA QRDR region (25).
3.5. Clinical Investigation
To address ATS recommendations regarding the treatment of M. simiae infected cases, a combination of levofloxacin (LEV), clarithromycin (CLR), and trimethoprim-sulfamethoxazole (TMP-SMX) was also clinically investigated (12, 26).
3.6. Statistical Analysis
The statistical analyses were performed using the Statistical Package for the Social Sciences (SPSS) (Version. 22.0; SPSS Inc. Chicago, IL, USA) software.
4. Results
4.1. Identification of Mycobacterium simiae and Subtype Classification
In this study, 7200 TB suspected specimens were referred to the National TB Reference Laboratory from May 2014 to May 2016. Only 240 clinical isolates were classified as NTM species, and the remaining 6960 were identified as M. tuberculosis complex. Culture results were also matched with the molecular examination.
As shown in Figure 1, after enzymatic digestion, PRA analysis identified a total of 60 M. simiae strains, and thus, the other species were excluded from the study. Surprisingly, M. simiae subtype I (Figure 1) was exclusively identified among M. simiae patients in Iran. In addition, the strains were detected in 35 (58.33%) and 25 (41.66%) males and females, respectively.
4.2. Drug Susceptibility Testing Against Mycobacterium simiae
A total of 60 M. simiae isolates were examined for susceptibility to RIF, INH, AMK, KAN, and CIP. Since all strains were identified as subtype I, the analysis of the responses of antibiotics to different subtypes of M. simiae was not applicable.
4.3. Susceptibility to Rifampin and Isoniazid
Rifampin and INH resistant cases were detected after fragment visualization on 8% polyacrylamide gel electrophoresis. As shown in Table 2, all M. simiae isolates were resistant to both RIF and INH. Therefore, resistance to the first-line drugs in the study population was 100%, while susceptibly to them was not demonstrated.
| Drug | Susceptible Isolates Recovered from | Susceptible Among All | |
|---|---|---|---|
| Male (n = 35) | Female (n = 25) | ||
| RIF | 0 | 0 | 0 |
| INH | 0 | 0 | |
| AMK | 31 (51.7) | 24 (40) | 55 (91.7) |
| KAN | 31 (51.7) | 24 (40) | 55 (91.7) |
| CIP | 31 (48.3) | 24 (40) | 53 (88.3) |
Frequency Proportion of Drug Susceptible Mycobacterium simiae in the Study a
4.4. Susceptibility to Amikacin and Kanamycin
Out of 60 M. simiae isolates, only 5 cases (8.33%) were resistant to AMK and KAN, while 55 isolates (91.66%) were susceptible (Table 2).
4.5. Susceptibility to Ciprofloxacin
Differentiation of patterns of M. simiae strains in contrast with wild-type patterns was implicated in mutations at gyrA. Compared to 7 (11.66%) resistant isolates, data analysis revealed the susceptibility of 53 cases (88.33%) to CIP (Table 2).
4.6. Clinical Results
Out of 60 M. simiae infected patients, clinical data for 58.33% (35/60) of the cases was available. Among them, 68.54% of the elderly (42.85% of males and 25.71% of females) were shown to be more prone to infection with the disease. In addition, susceptibility to M. simiae among middle-aged adults and young adults was 17.14% (8.57% of males and 8.57% of females) and 11.42% (8.57% of males and 2.85% of females), respectively. In contrast, the susceptibility among adolescents was 17.14% (8.57% of males and 8.57% of females). Noteworthy, children under the age of 11 and adolescent males were not susceptible to M. simiae infection. Also, 62.84% of the patients (37.14% of males and 8.14.27% of females) had a history of underlying diseases, and more than half of the patients (51.42%) had TB alone. Clinical evaluation revealed that only nine patients (25.71%; 14. 28% of males and 8.11.42% of females) were treated with a combination of LVX-CLR and TMP/SMX, although 11.42% of all cases (5.71% of males and 5.71% of females) did not receive therapy (unmonitored cases) and 5.71% of them (5.71% of males and 0% of females) are still undergoing treatment. Meanwhile, due to a lack of effective treatment response (28.57%; 8.57% of males and 20% of females), side effects (17.14%; 5.71% of males and 11.42% of females), and relapse after initial recovery (11.42%; 2.85% of males and 8.57% of females), the entire M. simiae infected cases (57.14%) failed treatment.
5. Discussion
After molecular examination, a total of 60 M. simiae patients were identified. Previously, Velayati et al. showed that M. simiae, with a frequency of 28.3%, was the most prevalent clinical NTM in Iran (10). Our result with a similar 25% detection ratio for M. simiae (compared to 240 NTM specimens) was consistent with their report. Heidarieh et al. noticed that out of 88 clinical slow-growing mycobacteria, M. simiae was detected in more than 50% of the cases (48 strains) (27). Also, Baghaei et al. demonstrated that out of 185 pulmonary patients referred to the National TB Reference Laboratory of Iran during 2002 - 2009, M. simiae was isolated from 26 cases (1). Among the other Middle Eastern countries, Turkey, Saudi Arabia, Oman, and Kuwait recently reported a few identified M. simiae cases. In contrast, the actual increasing rate of M. simiae infection in Lebanon raised to 47% (11, 28, 29). This highlights the notion that people with a particular ethnic origin, especially in the Middle East, are more prone to infection with M. simiae (28), indicating the importance of performing widespread research on these pathogenic mycobacteria.
Surprisingly, a total of 60 M. simiae strains were identified as subtype I. Since almost all mycobacterial specimens are referred to the National TB Reference Laboratory, subtype I may be confirmed as the most prevalent M. simiae subtype in Iran.
Recently, Hamieh et al. from Lebanon reported that males were predominantly (55%) infected with M. simiae (28). So, a higher proportion of males were infected with M. simiae in this study (58.33% males vs. 41.66% females). The World Health Organization (WHO) global report on the higher risk of TB in males could also be generalized into M. simiae infections in the Middle East and, in particular Iran (30). This may be associated with environmental, nutritional, and human genetic factors or host immunological response (31-33), but more gender-based investigations are necessary in this regard. Also, this study is the first to report that males and the elderly are more susceptible to infection with M. simiae in Iran.
Mycobacterium simiae strains are resistant to a wide range of recommended antibiotics for NTM treatment (12). Moreover, Heidarieh et al. illustrated that M. simiae strains in Iran were resistant to almost 80% of recommended mycobacterial antibiotics, which makes the selection of the most applicable regimen more elusive (27).
Given ATS recommendations (12), the susceptibility results also showed that M. simiae strains were resistant to both RIF and INH (100%). Hamieh et al. showed that M. simiae was 100% resistant to both RIF and INH, whereas Heidarieh et al. reported 77% resistant cases only to RIF (28). Thus, despite some similarities between M. tuberculosis and M. simiae, the first-line anti-TB drugs should be excluded from the treatment regimen of M. simiae patients.
Susceptibility of the cases to the second line anti-TB agents was completely in contrast. When both AMK and KAN showed 91.66% susceptibility, the sensitivity of the isolates to CIP was 88.33%. Only one patient was resistant to RIF, INH, AMK, KAN, and CIP. Compared to previous research with a high frequency of M. simiae, AMK was a far better selection with 88% susceptibility against M. simiae in Lebanon and more than half in Iran, but CIP was far less effective (100% resistant in Lebanon and 81% in Iran) (27, 28).
On the other hand, global reports on M. simiae treatment approaches have suggested the superiority of fluoroquinolones, including MOX, LEV, and CIP compared to the most common aminoglycosides (AMK and KAN), while our data demonstrated the higher susceptibility of M. simiae subtype I to AMK and KAN (12, 30). However, the slight difference between the susceptibility of the two groups of antibiotics (3.33%) may be ignored. This may be associated with differences between the performed methods to evaluate the susceptibility of the strains to antibiotics, different responses of M. simiae subtypes to the same antibiotic, and the studied countries or regions. Compared to the previous data from the Middle East, the latter is more reliable, where AMK revealed a better treatment response (27, 28). But an overall conclusion is not possible because of the following:
(1) No related study has been published to evaluate the association between different subtypes of M. simiae and the effective antibiotic regimen.
(2) Most of the previous studies on drug susceptibility of M. simiae have examined the minimal inhibitory concentration (MIC) using proportional methods (26).
(3) The limitation of M. simiae studies, particularly in the Middle East region, including Iran (11, 12, 27, 34).
In addition, combination therapy by LVX-CLR and TMP/SMX demonstrated that almost one-fourth of the patients (25.71%) were treated, and 57.14% of the cases failed treatment. Heidarieh et al. revealed that M. simiae isolates were resistant to CLR and TMP/SMX, as Hamieh et al. also showed 81% resistance to TMP/SMX but only 6% resistant to CLR (27, 28). Considering the same drug in use and the region, the far difference in CLR susceptibility among Iranian and Lebanese patients may be related to the different isolated M. simiae subtypes, but further subtyping-based research is needed. Altogether, due to the reliable susceptibility proportion of the second-line anti-TB agents in Iran, M. simiae patients could additionally receive AMK, and CIP may potentially replace LEV.
Comparing AMK and KAN, the global reports mostly recommended the use of AMK, while our data showed no differences between their susceptibility. Thus, regardless of the same study population and the drug susceptibility in vitro, and no differences in gender response to both antibiotics, AMK superiority in the treatment of M. simiae is more likely related to the specific isolated subtype (subtype I) or clinical-biochemical characteristics of the drug. The ATS recommendations and previous studies for treating patients infected with M. simiae somewhat confirm this theory (12-36).
Although ATS recommends using fluoroquinolones in the treatment of M. simiae (12), due to the resistance of some M. simiae strains to CIP, 100% reliability of CIP is not possible. On the other hand, ATS reported a better response of MOX/LEV in the majority of M. simiae cases, but as previously shown, for those Iranian M. simiae infected patients with suspected TB, who had been treated by the first-line anti-TB regimen, a combination of second-line therapy by CLR and CIP for two months or more saved them (37). So, highlighting the role of M. simiae subtypes in response to different fluoroquinolones, CIP showed better efficacy in treating M. simiae patients in Iran.
In conclusion, subtype I was exclusively identified among M. simiae patients in Iran. Molecular detection of drug resistance suggests that AMK/KAN, in conjunction with CIP, would likely comprise useful components of the antimicrobial drug regimen for patients infected with M. simiae subtype I.