1. Background
Coronavirus started a lethal pandemic in December 2019, which was named COVID-19 later (1). This disease was called “Severe Acute Respiratory Syndrome Coronavirus 2” (SARS-CoV-2), which imposed massive casualties and huge costs (2). Until October 2021, COVID-19 infected more than 5 million Iranians and caused 120,000 deaths (3).
Severe Acute Respiratory Syndrome Coronavirus 2 is a betacoronavirus with a positive-sense and single-stranded RNA enveloped with a protein capsid, especially M protein (4, 5). The genome of this virus, the same as SARS-CoV-1, consists of several structural and non-structural sequences enveloped with a set of accessory proteins responsible for virus replication and transmission (6). The ORF1ab gene encodes 16 non-structural proteins, such as RNA-dependent RNA polymerase (RdRp), helicase, 3’ to 5’ exonuclease, endoRNAse, and various proteases. The coronavirus structural proteins, including spike surface glycoprotein (S), envelope (E), membrane (M), and nucleocapsid (N), are expressed continuously. The ORF1ab/RdRp, E, N, and S genes are the most frequently used targets for COVID-19 detection in different RT-qPCR diagnostic kits (7, 8).
Real-time polymerase chain reaction (RT-PCR) is a method for detecting the presence of specific genetic material of any pathogen, including viruses. Therefore, RT-PCR is the most widely used laboratory method for detecting COVID-19 (9). Many countries have used RT-PCR for diagnosing COVID-19, but they have faced economic pressure to produce laboratory materials like filter columns for RNA extraction, especially in less developed and developing countries. Although some studies have reported that viral transport media (VTM) could be used directly in PCR reaction for SARS-COV-2 detection, this method also has some limitations related to the inhibitory effects of the sample (10, 11).
2. Objectives
in this study, we designed a simple and cost-benefit technique based on the isopropanol precipitation method to extract the viral genome from VTM without requiring a filter column and regular extraction kits. This technique is based on protein precipitation with ammonium acetate and nucleic acid precipitation with sodium acetate and isopropanol.
3. Methods
3.1. Sample Collection
The 41 nasopharyngeal swab samples of patients that were positive for COVID-19 were collected and stored at -80°C. All cases were positive for COVID-19 using an RT-PCR test and had symptoms like fever, sneezing, myalgia, and headache. All samples were collected with the patient’s consent and under the approved Jundishapur University of Medical Science ethical committee with the following registration code IR.AJUMS.REC.1399.955.
3.2. RNA Extraction with a Regular Extraction Kit (Column Base)
All samples were extracted using the viral RNA extraction kit (Rojeh, Iran) according to the manufacturer’s instructions. Briefly, 560 μL lysis buffer supplemented containing carrier RNA was added to 140 μL of sample and incubated for 10 min at room temperature. Then 560 μL of ethanol was added, vortexed, and transferred to the filter tube. After centrifugation, the filter column was washed with wash buffers 1 and 2. Finally, the pellet was dried and dissolved in 60 μL DEPC-treated water.
3.3. Nucleic Acid Extraction with the New Method (Precipitation Method)
After the samples were vortexed, 300 μL of VTM was added to a 1.5 mL microtube and put at 98 C for 5 min. The supernatant was transferred to a new microtube containing 45 μL of sodium acetate, 1 μL of polyacrylamide, and 500 μL of isopropanol. A new centrifugation step was made at 12000 rpm for 15 min at room temperature. The supernatant was discarded, and the pellet was washed with 70% ethanol to remove salts. The tube was uncovered for 5 minutes at a temperature range of 55°C to 60°C, letting the ethanol vaporize. Finally, the pellet was resuspended by 50 μL nuclease-free water to elute nucleic acid.
3.4. Nucleic Acid Extraction with Precipitation Method Without the Use of Ammonium Acetate
To evaluate the effect of ammonium acetate, all samples were heated at 98°C for 5 minutes. They were centrifuged at 12000 rpm for one min. Next, sodium acetate and ethanol were added and centrifuged at 12000 rpm for nucleic acid precipitation. Finally, the pellets were washed with the 70% ethanol and eluted in the elution buffer.
3.5. Real-time Polymerase Chain Reaction
All extracted samples were subjected to RT-PCR using the SARS-COV-2 RT-PCR kit (Pishtazteb, Iran) and MyGo Pro RT-PCR system (MyGo, UK). This kit uses RdRp and N regions of the virus genome to detect the virus, and RNase P is used as the internal control. 2.5 μL of samples extracted were added to a 10 μL RT-PCR reaction. If the results of Cts of RdRp, N, or both hit the limits, the patient’s sample would be considered positive.
4. Results
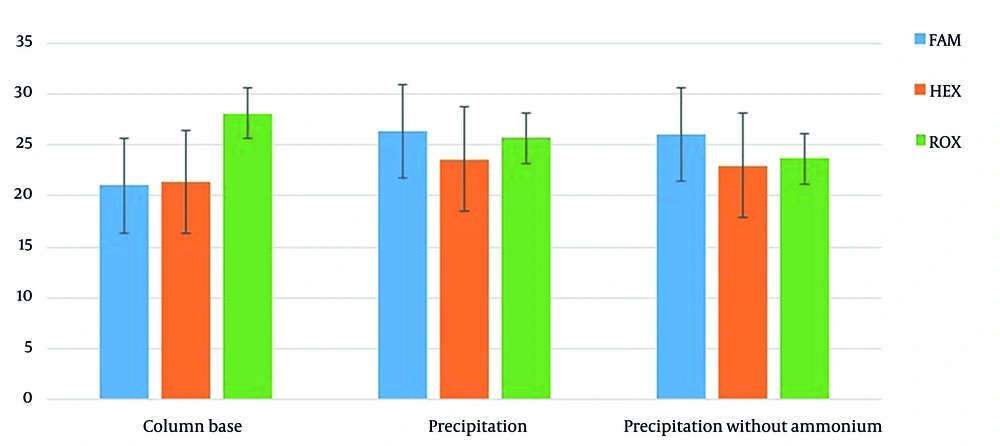
The 41 nasopharyngeal samples of positive COVID-19 patients were extracted with the new method based on precipitation, then compared with the conventional method (extraction kit by filter columns). The mean Cts in this group were 21 ± 4.9, 21.4 ± 4.8, and 28.1 ± 1.8 for the FAM channel (RdRp), HEX channel (N), and ROX (RNase P) channel, respectively. All the samples were positive (100%) for the N and RdRp genes in the column method. In the new extraction method, using ammonium acetate, sodium acetate, and ethanol, the average Ct was equal to FAM channel 26.3 ± 4.5, HEX channel 23.6 ± 5.3, and ROX channel (RNase P) 25.7 ± 3.5. The Ct values were close to the column base extraction method, especially in the HEX channel, where the differences between the two channels were not significant, P > 0.05, but The FAM channel Ct showed a significant difference, P < 0.05. In this extraction method, 40 out of 41 samples were positive (97.5%), and two (5%) were only positive for the N gene. This experiment was repeated without ammonium acetate, and again 40 out of 41 samples were positive (97.5%); but nine of them (22.5%) were only positive for the N gene without any signal in RdRp gene (FAM), which is about 4-fold more than extraction with ammonium acetate. Mean Cts were 26 ± 4.3, 23 ± 5.4, and 23.7 ± 2.3 for the FAM channel (RdRp), HEX channel (N), and ROX channel (RNase P), respectively, in this group (Figure 1). There was no significant difference in FAM and HEX channel Ct between extraction with and without ammonium acetate in the new extraction method, P > 0.05.
5. Discussion
The COVID-19 pandemic affected different countries worldwide, including health and the economy, especially in low-income countries. Economic side effects of COVID-19, such as prolonged lockdowns, loss of jobs, and the supportive role of governments in this situation, may put crushing pressures on developing countries (12). Diagnosis is one of the most expensive steps of crisis management. To extract and detect viral genes in nasopharyngeal swab samples a set of different extraction and detection kits are required (13). In this study, we utilized common chemicals to decrease costs as much as possible. Previous studies have indicated that adding VTM samples directly as templates to PCR reactions can be an inexpensive technique for detecting virus genomes (10, 11). However, there are two problems with this technique; first, the results are affected by the quality of different materials obtained from different manufacturers. The second is the sample’s purity. As a result of the use of direct samples in PCR reactions, the authors could only use 2l of VTM samples in a 20l PCR reaction to achieve the best results (10, 11, 14). To achieve a higher level of purity of samples, a simple and cost-benefit method was used to eliminate the inhibitors. Previous studies have suggested that high-speed centrifugation using the alcohol-free ammonium acetate solution may increase the purity of samples through the removal of proteins that play an inhibitory role in PCR (15). Our results demonstrated that the precipitation method by ammonium acetate has a Ct quantity nearly close to samples extracted by the column base method, especially in the HEX channel (N gene) and ROX channel (RNase P gene), however the differences between the two groups were seen in FAM channel (RdRp gene). The differences between the Cts of precipitation method by ammonium acetate extraction and the column-based in channel FAM (RdRp gene) could be due to the lower level of RdRp expression compared to the N gene, which may be more affected by the new extraction technique (16). As a result of the precipitation method, there were one false-negative result and two samples with undetectable FAM channels, while the column-based method yielded a higher purity. Generally, the precipitation method showed a 97.5% positivity rate in positive samples. For evaluation of the ammonium acetate effect in the extraction procedure, samples were extracted without ammonium acetate. In extraction without ammonium acetate, one false negative and nine undetectable FAM channels were found, which is more than four times higher than precipitation with ammonium acetate. There was no significant difference in Cts between extraction with and without ammonium acetate but regarding more false-negative and undetectable results in the extraction procedure without the ammonium acetate and the aforementioned limitations during the direct application of swab samples, it gives the impression that the use of ammonium acetate could result in much better detection of SARS-CoV-2 genes. These results lead us to two facts; the first one is that VTM has little inhibitory effect on the PCR reaction compared to complex specimens such as tissue or whole blood, which enables us to use it directly in PCR reaction in small volumes; and the second one is this fact that the titer of the virus in the nasopharyngeal swab of infected patients is relatively high as much as acceptable results were achieved despite using a small amount of it. However, the inhibitory effect of the VTM sample can still affect PCR results in higher volumes, especially when higher volumes of a target are needed in low titer samples. Polymerase chain reaction efficiency and reproducibility are also affected by the quality of materials, such as Taq polymerase and RT-PCR systems, when direct VTM samples are used as templates. However, the new procedure in this study can reduce the inhibitory effect of the VTM sample without the need for a column-based extraction kit.
5.1. Conclusions
Our results indicated that the precipitation method using ammonium acetate, sodium acetate, and ethanol could be an alternative extraction method instead of the column-based method for SARS-COV-2 by swab samples.