1. Background
Improper and uncontrolled antibiotic usage has led to the emergence of antibiotic resistance and superbugs. In recent years, global microbial resistance has significantly escalated (1). Concurrently, multidrug-resistant hospital-acquired infections (HAIs) are a leading cause of death in hospitalized patients (2). Hospital-acquired infections are typically not present before admission. These infections usually occur after hospitalization and manifest 48 hours after admission (3). Multidrug-resistant pathogens (MDRs) have become a major public health concern. Furthermore, antibiotic resistance results in prolonged hospital stays and escalates treatment expenses (4).
Patients admitted to the intensive care unit (ICU) are particularly vulnerable to HAIs due to underlying diseases, exposure to a variety of invasive procedures, heightened contact with health care personnel, and a long-term ICU stay (4). Infections in this unit constitute 25% of all HAIs, with incidence rates 5 to 7 times higher than those in general hospital wards (5). Globally, ICU discharge records show an average infection incidence of 9 per 100 patients (6). Antibiotic-resistant pathogens in ICU settings pose formidable challenges, rendering these infections increasingly difficult and, sometimes, impossible to treat (7). Epidemiological studies unequivocally demonstrate a direct link between antibiotic use and the emergence and dissemination of resistant strains in hospitals and ICUs (8). Studies indicate that 30 to 60% of the antibiotics prescribed in ICUs are unnecessary, inappropriate, or ineffective (9). Consequently, rational antibiotic administration and robust antibiotic stewardship practices are of paramount importance, especially given the emergence of superbugs and the paucity of new antibiotic development (10).
Effective management of antibiotic usage in ICUs entails swift identification and isolation of patients with bacterial infections, optimal selection of experimental treatment, utilization of pharmacokinetic-pharmacodynamic principles for dose optimization, and refined methods of antibiotic administration. These procedures could potentially reduce hospital length of stay. Studies have endorsed that comprehensive control of antibiotic administration and stewardship improves microbial susceptibility to antibiotics (11).
Antimicrobial susceptibility profiles of bacteria can substantially differ across geographic regions, hospitals, and even within different ICUs of the same hospital. Thus, to successfully implement rational antibiotic administration, a thorough understanding of local pathogen epidemiology and knowledge about evolving microorganism resistance patterns are imperative before selecting any drug regimen (12, 13).
2. Objectives
This study aimed to determine the frequency of bacteria isolated from patients with various infections in the ICU and to determine the pattern of antibiotic susceptibility, morbidity, and mortality rate of ICU patients in order to optimize the administration of antibiotic regimens, especially in MDR cases. This study was conducted in the largest teaching hospital in the Kurdistan Province of Iran, aiming to enhance antibiotic prescription guidelines.
3. Methods
3.1. Study Design
This cross-sectional study was conducted in the first 6 months of 2019 at Tohid Teaching Hospital in Sanandaj, Iran. This center has 518 beds, of which 33 beds belong to the ICU. The average monthly referral rate of patients from Iraq, surrounding cities, and other hospitals of Sanandaj to Tohid Hospital is 30. The average transfer rate of patients from other wards of the hospital to the ICU is 16 cases per month.
3.2. Clinical Setting and Data Collection
Sampling was performed on all the patients who were patients admitted to the ICU without any symptoms of infection but showed clinical signs of infection, including fever, tachycardia, tachypnea, respiratory distress, decreased blood pressure, increased endotracheal secretion, cellulite, phlebitis in intravenous (IV) line, and bedsores 48 hours after hospitalization. All the samples were taken under aseptic conditions. The information of the patients, including age, sex, number of days in the ICU, source of infection, culture results, isolated microorganisms, antibiogram, antibiotics prescribed, and treatment outcome (discharge or death), was collected using a checklist. The risk factors were assessed based on checklist variables.
3.3. Bacterial Isolation and Antimicrobial Susceptibility Testing
Clinical samples were cultured in general and specific culture media for 24 hours at 37°C, and if they did not grow, incubation was prolonged for another 24 hours. If the culture result was positive, the isolated bacteria were identified by standard methods, including Gram staining, colony characteristics, and finally, with the VITEK 2 device (bioMérieux, France). VITEK 2 was also used to evaluate antibiotic resistance. Special panels were used depending on the type of isolated bacteria, and minimum inhibitory concentration (MIC) results were recorded. Escherichia coli ATCC 25922 and Staphylococcus aureus ATCC 25923 were used as negative controls. The results were interpreted according to Clinical & Laboratory Standards Institute (CLSI) (14). Multi-drug resistance, extensive drug resistance (XDR), and pandrug resistance (PDR) definitions were determined (15).
3.4. Clonal Relatedness of Isolates
The repetitive extragenic palindromic-PCR (REP-PCR) molecular method was used to investigate the clonal relationship of the bacteria isolated from the ICU. In this method, deoxyribonucleic acid (DNA) was first extracted from fresh bacteria by the boiling method. Then, PCR was performed by REP1 and REP2 primers and a master mix (Pars Tous, Iran), as previously described (15). PCR products were run on 1.5% agarose gel and photographed. Images were embedded in GelJ v. 1.3, and the relevant dendrogram was extracted after adjusting the settings. Clones and clusters were determined with 98% and 90% similarity, respectively (16).
3.5. Statistical Analysis
The data collection was based on the census method, and the statistical analysis was performed in SPSS v. 25 (IBM, Chicago, IL, USA). Qualitative variables were calculated as frequency (percentage); quantitative variables with a normal distribution were calculated as mean and standard deviation (SD); and qualitative variables with an abnormal distribution were calculated as median (interquartile range (IQR)). A chi-square test was used to investigate the relationship between the variables and mortality. The P-value of 0.05 was considered as significant.
4. Results
4.1. Clinical and Demographical Analysis of Patients
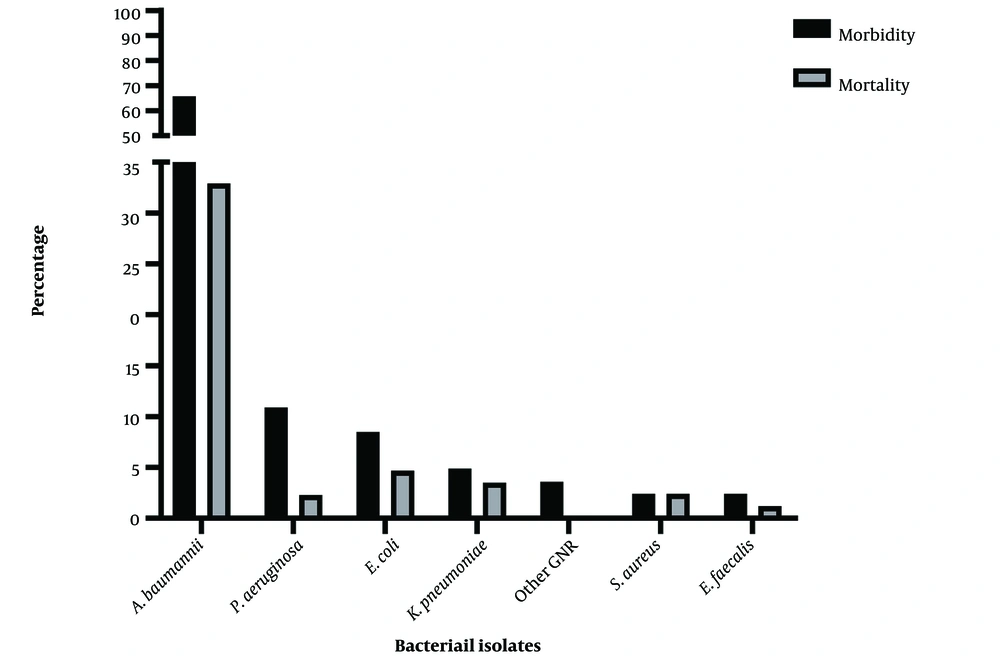
Of the 100 patients admitted to ICUs during the timeline of the study showing clinical signs of suspected nosocomial infection, 76 (76%) were positive; in some of them, two types of bacteria were isolated simultaneously, and 85 bacteria were collected in total (Table 1). Out of 76 patients from whom bacteria were isolated, 42 (55.3%) were female and 34 (44.7%) were male. The mean age of the patients was 64.9 ± 19.2, with a minimum and maximum of 23 and 90 years, respectively. The median length of IQR was 33 days in the ICU (15 - 60), and 43 patients (56.6%) died. The sources of infection included lung secretions (51.8%), urine (25.3%), blood (20.5%), and wounds (1.2%).
| Lung | Urine | Blood | Wound | Total | |
|---|---|---|---|---|---|
| Acinetobacter spp. | 30 (65.1) | 15 (65.2) | 9 (50) | 0 | 54 (63.5) |
| Pseudomonas aeruginosa | 4 (9.3) | 4 (17.4) | 0 | 1 (100) | 9 (10.6) |
| Escherichia coli | 5 (16.3) | 0 | 2 (11.1) | 0 | 7 (8.2) |
| Klebsiella pneumoniae | 3 (7) | 0 | 1 (5.5) | 0 | 4 (4.7) |
| Enterobacter cancerogenus | 1 (2.3) | 0 | 0 | 0 | 1 (1.2) |
| Enterobacter cloacae | 0 | 0 | 1 (5.5) | 0 | 1 (1.2) |
| Serratia marcescens | 0 | 0 | 1 (5.5) | 0 | 1 (1.2) |
| Staphylococcus aureus | 0 | 2 (8.7) | 2 (11.1) | 0 | 4 (4.7) |
| Enterococcus faecalis | 0 | 2 (8.7) | 2 (11.1) | 0 | 4 (4.7) |
| Total (n = 85) | 43 (100) | 23 (100) | 18 (100) | 1 (100) | 85 |
Frequency of Bacteria Isolated from Clinical Samples of Intensive Care Unit Patients and the Number of Deaths Among Patients with Hospital-Acquired Infections (%)
4.2. Bacterial Isolates
VITEK 2 results showed that out of 85 isolated bacteria, 78 were Gram-negative (90.6%), and only 8 (9.4%) were Gram-positive. Table 1 summarizes the frequency of different species of bacteria isolated from patients. The most common isolated microorganism was the Acinetobacter baumannii complex (63.5%), followed by Pseudomonas aeruginosa (10.6%) and E. coli (8.2%). The other Gram-negative bacteria were rare. For the first time in Iran, Enterobacter cancerogenus, which was resistant to carbapenems, was identified. Gram-positive bacteria isolated from the clinical specimens in this study included Staphylococcus aureus (4.7%) and Enterococcus faecalis (4.7%). The A. baumannii complex was the most common pathogen isolated in urine samples, lung secretions, and blood (Table 1).
4.3. Prescribed Antibiotics and Antimicrobial Resistance Profile
The most prescribed antibiotics were meropenem (71%), followed by vancomycin (56%) and ciprofloxacin (53%). The rate of other antibiotics was as follows: Levofloxacin (36%), amikacin (33%), ceftazidime (27%), clindamycin (26%), piperacillin/tazobactam (23%), ampicillin/sulbactam (21%), ceftriaxone (21%), colistin (20%), teicoplanin (18%), gentamicin (17%), cefepime (11%), imipenem (8%), and tobramycin (2%).
According to the antibiotic susceptibility testing (Table 2), colistin was the most effective antibiotic for Gram-negative bacteria, with only one isolate of the A. baumannii complex, one Klebsiella pneumoniae, and one Serratia marcescens being resistant. The other Gram-negatives were sensitive. Aminoglycosides were effective on P. aeruginosa (66.7% resistant) and E. coli (42.8% resistant) isolates. While all E. coli isolates were carbapenem-sensitive, all P. aeruginosa isolates were resistant (Table 2).
| Antibiotic | Organisms, No. (%) | |||||||
|---|---|---|---|---|---|---|---|---|
| Acinetobacter spp. (N = 54) | Escherichia coli (N = 7) | Pseudomonas aeruginosa (N = 9) | Klebsiella pneumoniae (N = 4) | Enterobacter spp. (N = 2) | Serratia marcescens (N = 1) | Enterococcus faecalis (N = 4) | Staphylococcus aureus (N = 4) | |
| Ticarcillin | NA | NA | 9 (100) | NA | NA | NA | NA | NA |
| Piperacillin | 54 (100) | 6 (85.7) | 9 (100) | NA | NA | NA | NA | NA |
| PIP/TAZ | 54 (100) | 6 (85.7) | 9 (100) | 2 (50) | 0 (0) | 1 (100) | NA | NA |
| Ceftazidime | 54 (100) | 6 (85.7) | 7 (77.7) | 2 (50) | 1 (50) | 1 (100) | NA | NA |
| Cefepime | 54 (100) | 6 (85.7) | 9 (100) | 2 (50) | 1 (50) | 1 (100) | NA | NA |
| Aztreonam | NA | 6 (85.7) | 8 (88.9) | 2 (50) | 0 (0) | 1 (100) | NA | NA |
| Imipenem | 54 (100) | 0 (0) | 9 (100) | 1 (25) | 1 (50) | 1 (100) | NA | NA |
| Meropenem | 54 (100) | 0 (0) | 9 (100) | 1 (25) | 1 (50) | 1 (100) | NA | NA |
| Colistin a | 1 (1.8) | 0 (0) | 0 (0) | 1 (25) | 0 (0) | 1 (100) | NA | NA |
| Ceftriaxone | 54 (100) | 0 (0) | NA | 2 (50) | 1 (50) | 1 (100) | NA | NA |
| Ertapenem | NA | 0 (100) | NA | 1 (25) | 1 (50) | 1 (100) | NA | NA |
| Cefuroxime | NA | ND | NA | 3 (75) | 3 (75) | 1 (100) | NA | NA |
| Amikacin | 52 (96) | 0 (0) | 6 (67) | 1 (25) | 0 (0) | 1 (100) | NA | NA |
| Tobramycin | 49 (90) | 3 (46.9) | 6 (3673) | 3 (75) | 1 (50) | 1 (100) | NA | NA |
| Gentamicin | 49 (90) | 3 (42.8) | 6 (67) | 2 (50) | 0 (0) | 1 (100) | NA | 3 (75) |
| Ciprofloxacin | 54 (100) | 6 (85.7) | 9 (100) | 2 (50) | 1 (50) | 1 (100) | 4 (0) | 2 (50) |
| Levofloxacin | 54 (100) | 6 (85.7) | 9 (100) | 2 (50) | 2 (100) | 1 (100) | 4 (0) | 2 (50) |
| Nitrofurantoin | NA | ND | NA | 3 (75) | 0 (0) | 1 (100) | 3 (75) | 0 (0) |
| SXT | 42 (77) | 0 (0) | NA | 1 (25) | 2 (100) | 0 (0) | NA | 0 (0) |
| Moxifloxacin | NA | NA | NA | NA | NA | NA | 4 (0) | 2 (50) |
| Erythromycin | NA | NA | NA | NA | NA | NA | 4 (100) | 3 (75) |
| Clindamycin | NA | NA | NA | NA | NA | NA | 4 (0) | 3 (75) |
| Linezolid b | NA | NA | NA | NA | NA | NA | 0 (0) | 0 (0) |
| Daptomycin | NA | NA | NA | NA | NA | NA | 0 (0) | 1 (25) |
| Vancomycin | NA | NA | NA | NA | NA | NA | 4 (100) | 0 (0) |
| Doxycycline | NA | NA | NA | NA | NA | NA | 3 (75) | 1 (25) |
| Tetracycline | NA | NA | NA | NA | NA | NA | 3 (75) | 1 (25) |
| Ampicillin | NA | NA | NA | NA | NA | NA | 3 (75) | NA |
| Oxacillin | NA | NA | NA | NA | NA | NA | NA | 1 (25) |
| Gentamicin high level | NA | NA | NA | NA | NA | NA | 4 (100) | NA |
| Streptomycin high level | NA | NA | NA | NA | NA | NA | 1 (25) | NA |
Frequency of Antibiotic Resistance for Pathogens Isolated from Patients Admitted to the Intensive Care Unit
The most effective antibiotic against Gram-positives was linezolid, with no resistance reported. In addition to the above antibiotic, vancomycin, nitrofurantoin, and cotrimoxazole had a 100% effect on S. aureus strains. One of the isolates in the cefoxitin screening was methicillin-resistant S. aureus (MRSA). All four E. faecalis isolates were resistant to vancomycin.
The multidrug resistance evaluation of the isolates showed that out of 85 bacterial species, 76 bacteria (89.4%) had the XDR phenotype, and 9 bacteria (10.6%) were MDR. Of all Gram-negative isolates, only one A. baumannii complex was PDR. All nine P. aeruginosa isolates and 9 isolates of Enterobacteriaceae had the XDR phenotype. Among the Gram-positive bacteria, 5 isolates (62%), 4 S. aureus, and all of E. faecalis were XDR.
4.4. Mortality and Risk Factors
Of the 76 patients from whom bacteria were isolated, 42 (55.2%) eventually died. The highest mortality rate in accordance with the species of bacteria was related to A. baumannii, in which 28 out of 42 (66.6%) patients expired. The mortality rate in other patients based on the isolated microorganisms is shown in Figure 1.
The mortality rate in patients with XDR infection was 95.2%, while that in MDR patients was 4.8%. There was a significant relationship between mortality and the emergence of XDR resistance (P = 0.034). The other risk factors were not significant (Table 3).
| Variables | Mortality, No. (%) | P-Value | |
|---|---|---|---|
| Yes (N = 42) | No (N = 33) | ||
| Age (y) | |||
| ≤ 70 | 19 (45.2) | 22 (66.7) | |
| > 70 | 23 (54.8) | 11 (33.3) | 0.10 |
| Sex | |||
| Female | 25 (59.5) | 16 (48.5) | |
| Male | 17 (40.5) | 17 (51.5) | 0.36 |
| Hospitalization (days) | |||
| ≤ 30 | 19 (45.2) | 14 (43.8) | |
| > 30 | 23 (54.8) | 18 (57.6) | 0.82 |
| Bacterial species | |||
| Acinetobacter | 25 (59.5) | 23 (69.7) | |
| Others | 17 (40.5) | 10 (30.3) | 0.47 |
| Resistance phenotype | |||
| XDR and PDR | 40 (95.2) | 27 (64.3) | |
| MDR | 2 (4.8) | 6 (35.7) | 0.034 |
Relationship of Mortality with Demographic and Clinical Characteristics of Patients
4.5. The Clonality Relationship of Isolates
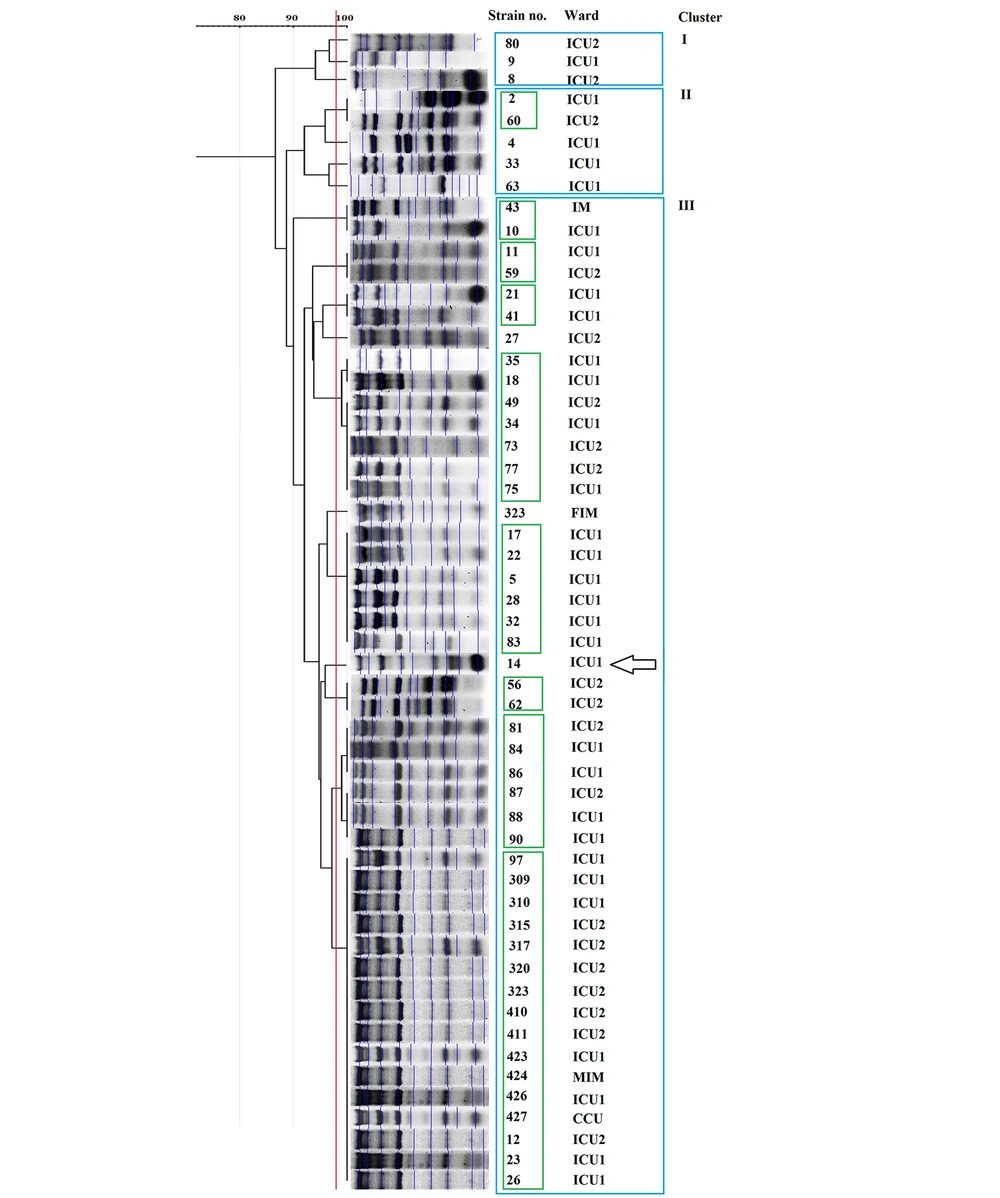
According to the REP-PCR results, there was limited genetic diversity among the isolates in the ICUs. Amongst them, 54 A. baumannii complex isolates belonged to 3 clusters (Figure 1, blue boxes), and 9 clones among the isolates. These results revealed that most of the species circulating between the ICUs of that hospital had a common genetic origin (Figure 1). The largest clone had 16 members, all of which were in cluster III. The REP-PCR technique discriminated the only colistin-resistant A. baumannii, Isolate 14, from other strains (Figure 2). The other bacteria were widely diverse, and no genetic link was found between them (data not shown).
5. Discussion
In this study, for the first time, risk factors and mortality rates associated with ICU patients and the genetic relationship among isolates were determined in the largest teaching hospital in Kurdistan province. Previous studies have shown that respiratory pathogens are responsible for approximately one-third of nosocomial infections in the ICU (17). In the present study, the most common site of infection was the respiratory tract (51.8%), consistent with the results of other studies in Iran (51.7%) (18), Turkey (38.8%) (19), Oman (65%) (20), and the United States (54.5%) (21). Numerous studies have tried to find the risk factors attributed to mortality in ICU patients (22, 23). The length of hospital stay, mechanical ventilation, and sedative usage before and after ICU hospitalization turned out to be risk factors for nosocomial infection in the ICU; meanwhile, the age over 60 years, acute physiology and chronic health evaluation II (APACHE II) score > 15, intubation, and central venous catheterization were significant risk factors for mortality.
S. aureus has been reported as the most common pathogen isolated from ICUs in developed countries, followed by P. aeruginosa and E. coli (21, 24). However, in developing countries such as Brazil, Turkey, Iran, and other Middle Eastern countries, P. aeruginosa and A. baumannii have been recorded as the most common pathogens (18, 20). In the present study, the A. baumannii complex was the most frequently isolated bacteria from clinical specimens in the ICU. Reports demonstrated the rising prevalence of A. baumannii in Asian countries and other continents (25). The mortality rate associated with this bacterium was estimated to be somewhere in the vicinity of 42.6% internationally (26); meanwhile, the results of our study showed that the mortality rate in our center was higher than the global average. The only mortality risk factor associated with this pathogen was its XDR phenotype, which was also mentioned in other studies (27). It was not possible to accurately estimate the mortality of the other isolated bacteria in this study due to the low number of isolates.
Several studies have reported a significant rise in A. baumannii resistance to common antibiotics, including colistin (28, 29). In a study by Salimizand et al., in a collection of A. baumannii isolates from this center from 2010 to 2012, 68% of the strains were resistant to carbapenems, while all the isolates in our 2019 study were carbapenem-resistant (30). This finding is significant since imipenem is commonly used to treat infections caused by A. baumannii; however, note that imipenem is currently ineffective. Therefore, colistin and cotrimoxazole are recommended as a combination therapy to treat carbapenem-resistant A. baumannii infections.
A VITEK 2 panel antibiotic is rifampicin, which has been used in the empirical treatment of PDR A. baumannii with promising results (31, 32). However, there is no established breakpoint in universal standards such as CLSI and European Committee on Antimicrobial Susceptibility Testing (EUCAST) for this antibiotic in Gram-negative bacteria. Therefore, it was impossible to interpret the results for A. baumannii isolates of our center. Gram-negative and Gram-positive bacteria are treated with these two antibiotics as a last resort. A combination therapy of two different classes of antibiotics is considered for isolates that are resistant to these last choices. Recent studies indicate that the mobilized colistin resistance gene (mcr) and its variants are significant causes of colistin resistance, which is spreading across bacteria due to the frequent use of colistin in veterinary medicine. The management of colistin-resistant Gram-negative bacteria that produce lactamase is therefore challenging. A new antibiotic called linezolid has been developed to treat drug-resistant Gram-positive bacteria, which belongs to the oxazolidinone class. Once the patient is clinically stable, it can be converted to oral therapy due to its high bioavailability. There are some Gram-positive infections for which it is effective, including certain drug-resistant strains of Enterococcus, Staphylococcus, and Pneumococcus. Due to the emergence of linezolid resistance, clinicians treating the infection will need to closely monitor antibiotic policies and antimicrobial stewardship programs.
The limitations of this study included the low number of clinical samples and the study of only one hospital. More samples are required to confirm the findings of the present study. Moreover, information regarding the follow-up of patients after hospital discharge will be valuable in finding the duration of HAI. The electronic infrastructure in our hospital setting lacked such information. Evaluation of antimicrobial resistance genes is another important issue that is valuable for epidemiological studies, but our facility could not prepare this information for this research.
5.1. Conclusions
The results of this study revealed a high mortality rate in Gram-negative isolates, especially A. baumannii. However, these findings must be confirmed by a larger sample. Moreover, A. baumannii strains in this center had high genetic similarities, indicating that the spread of this bacterium might be due to neglecting infection control preventive measures. Furthermore, the use of cotrimoxazole and colistin instead of carbapenems can be promising, especially in the treatment of infections caused by A. baumannii.