1. Background
Colorectal cancer (CRC) is the third most prevalent malignancy and the second leading cause of death globally (1). The global incidence of colon cancer is high and is rapidly increasing in developing countries (2). In Iran, CRC has emerged as one of the most common types of cancer, with a noticeable rise in recent years, as reported by the Iranian Cancer Society (3). Studies have indicated that CRC development or progression is influenced not only by genetic and lifestyle factors such as increased body mass index, obesity, reduced physical activity, and geographic location, but also by microbial and viral infections (4). Microbial agents disrupt intestinal homeostasis and can induce cellular and genetic alterations (5).
Among the various microbial agents, Helicobacter pylori infection is one of the most prevalent infections among the Iranian population (6). This bacterium has been classified as a class I carcinogen by the International Agency for Research on Cancer of the World Health Organization (7). The impact of H. pylori extends beyond gastric adenocarcinoma and has been associated with esophageal cancer, liver cancer, and CRC (8).
Campylobacter is recognized as one of the bacterial agents contributing to human digestive disorders (9). Moreover, this bacterium can cause asymptomatic infections in humans and act as a carrier state (10). It adheres to intestinal cells through CadF proteins and produces a genotoxin named cytolethal distending toxin (CDT), consisting of three protein subunits: CdtA, CdtB, and CdtC (11). Notably, the CdtB subunit possesses DNase I properties, which can induce damage to the host's DNA, leading to mutations in intestinal cells and promoting the development of CRC (12).
In addition, viral agents with carcinogenic properties contribute to approximately 10 - 20% of all cancer cases (13). Viral infections, particularly human papillomavirus (HPV) and JC polyomavirus (JCV), can also have a significant impact on CRC development (14, 15). HPV has been associated with various malignancies, including cervical cancer and lung cancer (16, 17). Recent studies have shown the role of HPV in CRC, but the findings have been contradictory (18). On the other hand, JCV, a member of the polyomavirus family, has been identified as a potential factor in the development of lower gastrointestinal neoplasms, including CRC (13).
The impact of bacterial and viral infections on the onset and progression of CRC in the Iranian population remains uncertain and controversial.
2. Objectives
Therefore, in the present study, the prevalence of a selection of gastrointestinal microbiota, including H. pylori, different species of Campylobacter, HPV, and JCV, in tissue samples of Iranian CRC patients was analyzed using the polymerase chain reaction (PCR) method.
3. Methods
3.1. Human Tissue Sample Collection
In this cross-sectional descriptive study, a total of 86 colon biopsy specimens, including 17 samples from healthy individuals and 69 from CRC patients, were collected at Taleghani Hospital in Tehran, Iran, between 2017 and 2019. A questionnaire was used to record demographic information, medical history, and details regarding physical activity, such as walking and running, prior to colonoscopy. Patients with complex diseases such as diabetes or high blood pressure, a history of cancer, inflammatory bowel disease, or those who had taken antibiotics or non-steroidal anti-inflammatory drugs within the 3 months prior to tissue sampling were excluded.
Suspected CRC patients were identified based on evaluations by expert gastroenterologists, considering the presence of suspicious clinical symptoms related to the disease. Examples of such symptoms include rectal bleeding, blood in the stool, frequent abdominal discomfort such as cramping and bloating, family history, age, and other related factors. Diagnosis of suspected CRC patients was confirmed through colonoscopy and histopathological examination. Normal controls consisted of participants with no abnormal colonoscopy results and a negative family and personal history of gastrointestinal diseases.
For all patients, at least three biopsy specimens were taken. Two samples were sent for histopathological examination, while one sample was used for molecular tests. Biopsy specimens were carefully placed in sterile screw-top test tubes and promptly transported to the laboratory, where they were stored at -80°C for further analysis. This study was approved by the ethics committee of Shahid Beheshti University of Medical Sciences, and informed consent was obtained from all patients, indicating their willingness to participate in the study.
3.2. Bacterial Species and Target Genes
The reference strains, including H. pylori CCUG 17874, Campylobacter coli ATCC 33559, and Campylobacter jejuni ATCC 33560, were obtained from the Foodborne and Waterborne Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran. Campylobacter fetus ATCC 33561, Campylobacter lari ATCC 35221, and Campylobacter upsaliensis ATCC 43954, as well as the control DNA samples of HPV and JCV, were provided by Tarbiat Modares University, Tehran, Iran. The bacterial reference strains were cultured on 10% Columbia blood agar (Merck Co., Hamburg, Germany) and incubated at 37°C under microaerobic conditions (86% N2, 5% O2, 5% CO2, and 4% H2) for 3 - 5 days.
3.3. Total DNA Extraction
Genomic DNA was extracted from tissue samples and harvested colonies of reference strains using a commercially available DNA Extraction Kit (QIAamp DNA Mini, USA) according to the manufacturer's instructions. The quality of the extracted DNA was assessed by measuring the absorbance at 260 and 280 nm using a Nanodrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The samples were also subjected to electrophoresis on 2% agarose gels. All DNA samples were then stored at -20°C until further use.
3.4. PCR Amplification
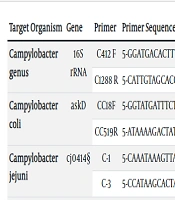
The presence of bacterial and viral agents was determined using PCR with specific primers. All primer pairs were evaluated using the NCBI BLAST tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to ensure their specificity. A no-template reaction mixture was used as a negative control. All PCR reactions were carried out in 0.2 mL tubes using an Eppendorf thermal cycler. Each PCR reaction consisted of a 25 µL solution containing 300 ng of DNA template, 1.0 µM of each primer, 0.25 U of Taq DNA polymerase, 0.5 mM dNTPs, and 1 mM MgCl2. The PCR conditions included an initial denaturation step at 95°C for 5 minutes, followed by 34 cycles of denaturation at 95°C for 45 seconds, annealing at 56°C for 40 seconds, and extension at 72°C for 1 minute. A final extension step at 72°C for 5 minutes was performed. The oligonucleotide sequences used in this work are listed in Table 1.
| Target Organism | Gene | Primer | Primer Sequence | Product Size (bp) | Reference |
|---|---|---|---|---|---|
| Campylobacter genus | 16S rRNA | C412 F | 5-GGATGACACTTTTCGGAGC-3 | 816 | (19) |
| C1288 R | 5-CATTGTAGCACGTGTGTC-3 | ||||
| Campylobacter coli | askD | CC18F | 5-GGTATGATTTCTACAAAGCGAG-3 | 502 | (20) |
| CC519R | 5-ATAAAAGACTATCGTCGCGTG-3 | ||||
| Campylobacter jejuni | cj0414 | C-1 | 5-CAAATAAAGTTAGAGGTAGAATGT-3 | 161 | (20) |
| C-3 | 5-CCATAAGCACTAGCTAGCTGAT-3 | ||||
| Campylobacter fetus | cstA | MG3F | 5-GGTAGCCGCAGCTGCTAAGAT-3 | 359 | (20) |
| CF359R | 5-AGCCAGTAACGCATATTATAGTAG-3 | ||||
| Campylobacter lari | glyA | CLF | 5-TAGAGAGATAGCAAAAGAGA-3 | 251 | (20) |
| CLR | 5-TACACATAATAATCCCACCC-3 | ||||
| Campylobacter upsaliensis | lpxA | CU61F | 5-CGATGATGTGCAAATTGAAGC-3 | 86 | (20) |
| CU146R | 5-TTCTAGCCCCTTGCTTGATG-3 | ||||
| Helicobacter genus | 16S rRNA | H276f | 5-CTATGACGGGTATCCGGC-3 | 357 | (21) |
| H676r | 5-ATTCCACCTACCTCTCCCA-3 | ||||
| Helicobacter pylori | glmM | glmM-F | 5-GCTTACTTTCTA ACACTAACGCGC-3 | 296 | (22) |
| glmM-R | 5-GGATAAGCTTTTAGGGGTGTTAGGGG-3 | ||||
| JCV | VP | P-F | 5-AGGAGGTGCAAATCAAAGATCTG-3 | 102 | (23) |
| P-R | 5-GGGCCATCTTCATATGCTTCA-3 | ||||
| HPV | L1 | GP5 | 5-TTTGTTACTGTGGTAGATAC-3 | 155 | (24) |
| GP6 | 5-GAAAAATAAACTGTAAATCA-3 |
3.5. Data Analysis
Chi-squared and Fisher's exact tests were employed for the analysis of categorical data, while the t-test was used for numerical data. The statistical analyses were performed using SPSS V22 (SPSS, Chicago, IL, USA). A P-value of < 0.05 was considered statistically significant.
4. Results
The demographic and clinical variables of the participants in different study groups were analyzed, and their P-values were assessed between the normal and CRC groups. The mean age of the patients with cancer was 56.85 ± 16 years, while the mean age of the normal subjects was 54.19 ± 14.17 years. The difference in mean age between the two groups was not statistically significant (P = 0.34).
Regarding gender distribution, 46 (66.7%) of the cancer patients and 12 (70%) of the normal subjects were male. There was no significant difference in gender distribution between the two groups (P > 0.05). Additionally, no significant differences were found between the two groups considering factors such as family history of CRC, history of diabetes, smoking, and exercise (P > 0.05) (Table 2).
| Characteristics | Normal (N = 17) | Cancer (N = 69) | Total (N = 86) | P-Value |
|---|---|---|---|---|
| Age | 54.19 ± 14.17 | 56.85 ± 16.00 | 56.35 ± 15.62 | 0.348 |
| Sex | 1 | |||
| Male | 12 (70) | 46 (66.7) | 54 (62.8) | |
| Female | 5 (30) | 23 (33.3) | 28 (32.6) | |
| Family history of CRC | 0.45 | |||
| Yes | 1 (5.9) | 12 (17.4) | 13 (15.1) | |
| No | 16 (94.1) | 57 (82.6) | 73 (84.9) | |
| Diabetes | 0.37 | |||
| Yes | 3 (17.6) | 6 (8.7) | 9 (10.5) | |
| No | 14 (82.4) | 63 (91.3) | 77 (89.5) | |
| Alcohol consumption | 0.33 | |||
| Yes | 2 (12) | 4 (6) | 6 (1.2) | |
| No | 15 (88) | 65 (94.6) | 80 (94.2) | |
| Smoking | 0.4 | |||
| Yes | 3 (20) | 7 (8.7) | 10 (8.1) | |
| No | 14 (80) | 62 (91.3) | 76 (91.9) | |
| Exercise | 0.72 | |||
| Yes | 2 (12) | 14 (20) | 16 (23.3) | |
| No | 15 (88) | 55 (80) | 70 (76.7) | |
| Anatomical site of biopsy | 0.10 | |||
| Cecum | 2 (11.7) | 4 (5.8) | 6 (7.0) | |
| Rectum | 6 (35.3) | 22 (31.9) | 28 (32.5) | |
| Descending colon | 2 (11.7) | 11 (15.9) | 13 (15.1) | |
| Rectal | 1 (5.9) | 2 (2.9) | 3 (3.5) | |
| Rectosigmoid junction | 1 (5.9) | 12 (17.4) | 13 (15.1) | |
| Colon | 1 (5.9) | 12 (17.4) | 13 (15.1) | |
| Transverse colon | 2 (11.7) | 1 (1.4) | 3 (3.5) | |
| Anus mass | 1 (5.9) | 1 (1.4) | 2 (2.3) | |
| Ascending colon | 1 (5.9) | 4 (5.7) | 5 (5.8) | |
| Total | 17 (100) | 69 (100) | 86 (100) |
a Values are expressed as No. (%) or Mean ± SD.
In terms of the prevalence of the studied bacteria in both groups, 14 (20.3%) of the cancer patients and 2 (12%) of the normal subjects tested positive for the Campylobacter genus. Among the positive cases for Campylobacter, 6 (8.7%) were identified as C. coli, 5 (7.2%) as C. upsaliensis, and 3 (4.3%) as C. lari. The prevalence of the Helicobacter genus and HPV was observed in 27 (39.1%) and 29 (42.0%) of the cancer patients, respectively. Additionally, 33 (47.0%) of the cancer patients and 2 (11.8%) of the control subjects tested positive for JCV (Table 3).
| Target organism | Normal (N = 17) | Cancer (n = 69) | Total (n = 86) | P-Value |
|---|---|---|---|---|
| Campylobacter genus | 0.032 | |||
| Yes | 2 (12) | 14 (20.3) | 16 (19) | |
| No | 15 (88) | 55 (79.7) | 70 (81) | |
| Campylobacter coli | 0.594 | |||
| Yes | 0 (0) | 6 (8.7) | 6 (7.0) | |
| No | 17 (100.0) | 63 (91.3) | 80 (93.0) | |
| Campylobacter jejuni | 1 | |||
| Yes | 0 (0) | 0 (0) | 0 (0) | |
| No | 17 (100.0) | 69 (100.00) | 69 (80.2) | |
| Campylobacter fetus | 1 | |||
| Yes | 0 (0) | 0 (0) | 0 (0) | |
| No | 17 (100.0) | 69 (100.00) | 86 (100.0) | |
| Campylobacter lari | 0.610 | |||
| Yes | 0 (0) | 3 (4.3) | 3 (3.5) | |
| No | 17 (100.0) | 66 (95.7) | 83 (96.5) | |
| Campylobacter upsalinsis | 0.578 | |||
| Yes | 0 (0) | 5 (7.2) | 5 (5.8) | |
| No | 17 (100.0) | 64 (92.8) | 81 (94.2) | |
| Helicobacter genus | 0.001 | |||
| Yes | 0 | 27 (39.1) | 27 (31.4) | |
| No | 17 (100.0) | 42 (60.9) | 59 (68.6) | |
| H. pylori | 1 | |||
| Yes | 0 (0) | 0 (0) | 0 (0) | |
| No | 17 (100.0) | 69 (100.0) | 86 (100.0) | |
| JCV | 0.006 | |||
| Yes | 2 (11.8) | 33 (47.8) | 35 (40.7) | |
| No | 15 (88.2) | 36 (52.2) | 51 (59.3) | |
| HPV | 0.001 | |||
| Yes | 0 | 29 (42.0) | 29 (33.7) | |
| No | 17 (100.0) | 40 (58.0) | 57 (66.3) |
a The abundance of different microorganisms in each group. P-value less than 0.05 was considered statistically significant.
b Values are expressed as No. (%).
Interestingly, the prevalence of the Campylobacter genus, Helicobacter genus, HPV, and JCV was significantly higher in patients with CRC compared to the normal controls (P < 0.05). However, there were no statistically significant differences in the rates of other studied bacteria, including C. coli, C. upsaliensis, and C. lari, between the two groups (P > 0.05) (Table 3).
5. Discussion
The human gut hosts a diverse community of microbes that vary in type and abundance across different regions of the intestine (25). Recent research has revealed that an imbalance in the gut microbiota, known as dysbiosis, is associated with the progression of inflammatory bowel disease to CRC (26). Evidence suggests that microbial agents, such as bacteria and viruses, can significantly affect various processes in CRC development, including DNA damage, induction of inflammation, and production of proinflammatory cytokines and interleukins (27, 28). However, it is important to note that the exact mechanisms and causal relationships between host cells and the gut microbiota have not been definitively elucidated (29, 30).
This study aimed to investigate the relationship between the prevalence of gastrointestinal microbiota, including H. pylori, different species of Campylobacter, HPV, and JCV in CRC biopsy samples. In our study, H. pylori was not found in cancer tissue samples, but the Helicobacter genus was detected at a prevalence of 39.1%, which could be related to other Helicobacter species. The exact mechanism by which H. pylori contributes to the progression of CRC is a subject of debate. Non-pylori Helicobacter species such as H. canis, H. marmotae, and H. bilis are a group of opportunistic microorganisms that can stimulate host inflammatory cascades and lead to genetic or epigenetic changes (31, 32).
In a study by Bulajic et al., PCR amplification was used to investigate the prevalence of H. pylori in CRC tissue samples (33). In contrast to our study, these researchers detected H. pylori DNA in 1.2% of CRC patients and 6% of normal colonic mucosa samples. However, they found no significant association between H. pylori and CRC. In another study, Liou et al. identified H. pylori infection in 3.6% of CRC and 21.4% of normal mucosa samples in Taiwan (34). Similarly, they found no significant relationship between H. pylori infection and an increased risk of CRC.
Campylobacter species are considered endemic in developing countries such as Iran, and infected individuals are often asymptomatic carriers (35). Currently, limited studies have investigated the role of Campylobacter species in the development of CRC. However, previous studies have demonstrated an association between CRC and cytolethal distending toxin (Cdt) produced by Campylobacter species. Cdt can induce apoptosis and autophagy signaling pathways, leading to increased inflammation and chromosomal instability (36, 37). A study by Pickett and Whitehouse demonstrated that low doses of Cdt (50 pg/mL) can induce early DNA damage and DNA double-strand breaks (DSBs), leading to prolonged arrest in the cell cycle in the G1 and/or G2 phase (38). This event resulted in the production of inflammatory cytokines, promoting tumor progression and metastasis (38). According to our results, the prevalence of the Campylobacter genus was 20.3% in CRC patients and 12% in the normal control group. Statistical analysis showed a significant association between the prevalence of Campylobacter in CRC tissues compared to normal controls (P < 0.05). These findings are consistent with other studies showing a significant association between Campylobacter infection and colon malignancies (10, 39, 40). However, further research is required to elucidate the potential role of Campylobacter infection in the development of CRC.
A considerable number of studies have reported a direct link between high-risk papillomavirus (HPV) infection and CRC (18, 41). A recent study in Taiwan showed a high frequency of HPV infection in CRC patients (42). The data from Damin's study revealed the presence of HPV in patients diagnosed with CRC, suggesting a potential association between this virus and the development of CRC (18). Our research, consistent with other studies, demonstrated a high prevalence of HPV infection among patients with CRC (42.0% in CRC vs. 0% in controls). These findings suggest a potential involvement of HPV in the development of CRC in the Iranian population.
Limited studies have been conducted on the status of JCV in CRC patients. Laghi et al. found that JCV DNA was present in the mucosa of the human colon and CRC, suggesting the virus's involvement in colon cancer pathogenesis and tumor development (43). In our study, JCV was detected in almost half (48%) of CRC patients compared to 12% in the normal control, indicating a potential role of JCV in CRC development among Iranian patients (P < 0.05). Similar to our findings, a study by Lin explored the relationship between CRC and JCV, revealing the presence of virus DNA in CRC patients and a high prevalence of JCV infection in colon cancer tissue in Taiwan (44)). Unlike our observations, Karbalaie Niya et al. reported a low prevalence of JCV infection in the Iranian CRC population (45). However, more research is needed to investigate the pathogenic role of these viruses in CRC development and progression.
5.1. Conclusions
In conclusion, our study demonstrated a higher presence of the Helicobacter genus, Campylobacter species, as well as HPV and JCV, in colorectal specimens from patients with CRC compared to the normal control group. This supports the potential role of these organisms in CRC development among Iranian patients. However, we did not detect H. pylori in either patients with CRC or the normal control group, suggesting no correlation between H. pylori and CRC development in Iranian patients. It is important to note that our study was conducted with a limited number of samples; therefore, further research with a larger sample size and population is recommended for more precise conclusions.