1. Background
Staphylococcus aureus is a naturally occurring gram-positive coccus and a common cause of healthcare-associated infections and community-acquired pathogens (1). Nowadays, treating staphylococcal infections has become increasingly difficult due to the acquisition of antibiotic resistance factors, so much so that with the introduction of each new antibiotic, resistant bacterial strains have rapidly emerged (2). The treatment of staphylococcal infections by beta-lactam drugs was very successful until 1942 (3). However, the first identification of methicillin-resistant S. aureus (MRSA) containing the mecA gene marked the beginning of resistance to common antibiotic treatments (4). Studies have shown that about half of the S. aureus isolates from ICUs in European countries are resistant to methicillin (5).
Since beta-lactam antibiotics are no longer effective in treating MRSA isolates, glycopeptides are now used as the main replacement drug of choice for treating staphylococcal infections. The use of glycopeptide antibiotics, such as vancomycin, has increased over the last two decades due to the rising prevalence of MRSA and methicillin-resistant coagulase-negative staphylococci (6). MRSA infection is reported to be one of the leading infectious diseases globally, due to its high rates of morbidity and mortality, posing a serious threat to human health and drawing the attention of the global medical community. Therefore, it is urgent to monitor the genes involved in resistance mechanisms (7).
To date, the genetic origins associated with vancomycin-resistant S. aureus (VRSA) are clear, and a molecular biological method for the detection of VRSA strains is also available. The van genes are especially important in the development of resistance to vancomycin. The vanA and vanB genes are associated with high resistance to vancomycin. Other genes of this family, including vanC, D, E, G, L, located on chromosomes, can result in a lower level of resistance. The relatively high level of vanA and vanB in VRSA strains indicates a high potential for horizontal gene transfer from other vancomycin-resistant species or one of the other vanA and vanB positive bacteria to S. aureus (1). The other vanA and vanB negative VRSAs suggest that possible changes in cell membrane such as low permeability as well as thickened and poorly cross-linked cell wall may be responsible for the presence of vancomycin resistance.
Type A and B are van genes located on Tn-1546 and Tn-1549 transposons, respectively. Transposon 1546 is a 10851 base sequence that is generally found within the plasmid PLw1043 and vancomycin-susceptible strains lack this transposon. The plasmid can transmit through conjugation between different Enterococcus also between different genera of bacteria including S. aureus, which has acquired vancomycin resistance in this way (8, 9).
With the development of vancomycin resistance, the need for replacement drugs has been identified in the community. Since 2000, the oxazolidinone class of antibiotics, including linezolid, has been used to treat vancomycin-resistant enterococcus infections as well as MRSA and VRSA (10).
While linezolid is a comparatively new antibiotic, resistance to this drug has been observed in various countries (11). One of the mechanisms of linezolid resistance is the presence of the cfr gene, which was found in 1997 on a plasmid called pSCFS1 in S. sciuri isolated from a calf (12). The cfr gene appears to have been transmitted horizontally among Staphylococcus strains. Although the presence of cfr only slightly enhances linezolid MIC in the absence of ribosomal mutations, its ability to spread among different strains or species is a worldwide concern (13).
2. Objectives
The aim of this study was to identify the molecular characteristics of vancomycin and linezolid resistance by detecting the frequency of VRSA and VISA strains and van genes. Additionally, the study aimed to investigate the frequency of resistance to linezolid and the responsible gene (cfr) in clinical isolates of methicillin-resistant S. aureus collected from Qazvin and Tehran hospitals in Iran. This information is intended to establish a resistance profile for vancomycin and linezolid to develop appropriate treatment and infection control protocols.
3. Methods
3.1. Clinical Isolates
In this descriptive epidemiological study, specimens were collected from patients admitted to educational hospitals in Qazvin and Tehran over a four-year period (2014 - 2018). The samples were investigated in five hospital laboratories for the identification of S. aureus using biochemical tests. Samples were collected using a simple random method. Patient demographics and clinical information were extracted from their medical records. The inclusion criteria were all samples of Staphylococcus aureus isolated from inpatients in the ICU and other wards, while all outpatient and repeat samples were excluded. All samples were initially cultured and isolated into nutrient agar for purity. The isolates were identified using biochemical and molecular techniques (femA amplification). A total of 270 isolates of S. aureus were identified and cultured in sterile TSB medium containing 30% glycerol and blood for storage at -80°C for further study (14).
3.2. Antibiotic Susceptibility Testing and Detection of MRSA
The antibiotic susceptibility of the isolates was determined using the disk diffusion method and the Kirby Bauer assay according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (15). The selected antibiotic disks for this assay included cefoxitin (30 μg), sulfamethoxazole-trimethoprim (1.25/23.7 μg), clindamycin (2 μg), gentamycin (15 μg), tetracycline (30 μg), and ciprofloxacin (5 μg). The S. aureus ATCC 25923 standard strain was used as the quality control for the disks, which included cefoxitin (30 μg) and linezolid (30 μg) (14).
3.3. Minimum Inhibitory Concentration of Vancomycin
The minimum inhibitory concentration (MIC) of vancomycin was determined using agar dilution and E-test methods. The agar dilution MIC was performed according to the 2018 CLSI guidelines. S. aureus ATCC29213 was used as a positive control, as specified by the CLSI Quality Control Table. The results for vancomycin-non-susceptible isolates were interpreted based on CLSI standards. The E-test was conducted on Mueller Hinton agar, with the standard strain ATCC29213 serving as the positive control (16).
3.4. Genomic DNA Extraction
Total DNA extraction was carried out using a commercial kit (DNA Extraction Kit, Sinaclon, CinnaGen Inc, Iran) following the manufacturer’s instructions, with some modifications and additional preparation steps. The DNA concentration was measured in micrograms per milliliter using A260 values obtained from a Nanodrop system (ABI, USA).
3.5. Detection of Resistance Genes by PCR and Sequencing
PCR was used to amplify the mecA, mecC, vanA, vanB, vanC, and cfr genes to detect methicillin, vancomycin, and linezolid resistance, respectively. The specific primers used are listed in Table 1. The PCR protocol included an initial denaturation at 94°C for 2 minutes, followed by 30 cycles of 94°C for 10 seconds, 59°C for 15 seconds, and 72°C for 30 seconds, with a final extension at 72°C for 5 minutes. The PCR products were then electrophoresed on a 1% agarose gel. Sequencing of PCR products was performed using a genetic analyzer (Applied Biosystems, USA), and the resulting data were analyzed using NCBI tools.
| Gene | Primer | Sequence: 5'-3' | Amplicon, bp | Ref. |
|---|---|---|---|---|
| mecA | mecA-F | AACAGGTGAATTATTAGCACTTGTAAG | 171 | (17) |
| mecA-R | ATTGCTGTTAATAATTTTTGAGTTGAA | |||
| mecC | mecC-F | TGTTGTAGCAATGTTCACAC | 138 | (18) |
| mecC-R | CAAGCACTTAATATCAACGC | |||
| vanA | VanA- F | ATCAACCATGTTGATGTAGC | 136 | (19) |
| VanA- R | AAGGGATACCGGACAATTCA | |||
| vanB | VanB- F | ACCCTGTCTTTGTGAAG | 121 | (19) |
| VanB-R | GAAATCGCTTGCTCAAT | |||
| vanC | VanC-F | CAGCAAGTGTGATCCAAGC | 300 | Were designed |
| VanC-R | CGACATGGCAACCAACATAAG | |||
| cfr | Cfr-F | TGAAGTATAAAGCAGGTTGGGAGTCA | 746 | (20) |
| Cfr-R | ACCATATAATTGACCACAAGCAGC | |||
| femA | femA-F | AAAAAAGCACATAACAAGCG | 132 | (21) |
| femA-R | GATAAAGAAGAAACCAGCAG |
3.6. Data Analysis
The data were analyzed using SPSS v.21 software with chi-Square and Fisher tests (95% confidence interval). A P-value < 0.05 was considered statistically significant.
4. Results
4.1. Demographic Characteristics
A total of 270 isolates were collected from teaching hospitals in Qazvin and Tehran. Of the 270 patients admitted to the hospitals, 161 (59.6%) were male and 109 (40.4%) were female. The mean age of the hospitalized patients was 44.8 years. S. aureus isolates were obtained from various samples, primarily from blood (23.3%), sputum (20%) and wound (18.1%), with fewer from tissue (0.7%) and nose (0.7%) specimens (Table 2). Most isolates were collected from internal units (40.7%) and intensive care units (27%).
| Sample | No. (%) | Hospital Ward | No. (%) |
|---|---|---|---|
| Blood | 63 (23.3) | Internal | 110 (40.7) |
| Sputum | 54 (20) | ICU | 73 (27) |
| Wound | 49 (18.1) | Infectious | 40 (14.8) |
| Catheter | 27 (10) | Neonatal | 19 (7) |
| Urine | 24 (8.9) | Surgery | 10 (3.7) |
| Abscess | 19 (7) | Neurology | 4 (1.4) |
| Respiratory | 22 (8.1) | Orthopedy | 4 (1.4) |
| Nose | 2 (0.7) | CCU | 3 (1.1) |
| Boil | 4 (1.5) | Gastroenterology | 2 (0.7) |
| Tissue | 2 (0.7) | Burn | 2 (0.7) |
| Eye | 4 (1.5) | Ophthalmology | 2 (0.7) |
| Total | 270 (100) | Radiotherapy | 1 (0.4) |
4.2. Antibiotic Susceptibility Testing and Detection of MRSA
Antibiotic susceptibility for linezolid and cefoxitin was determined using the agar disk diffusion method in accordance with CLSI guidelines (CLSI 2018). Resistance to linezolid was observed in 10 (3.7%) isolates. Additionally, 152 (56.3%) isolates were resistant to cefoxitin and were classified as methicillin-resistant S. aureus (MRSA). The most effective antibiotics against S. aureus were sulfamethoxazole-trimethoprim (82.6%), clindamycin (58.2%), and gentamycin (51.7%), while the highest resistance rates were detected for tetracycline (58.3%) and ciprofloxacin (50%).
4.3. Minimal Inhibitory Concentration of Vancomycin
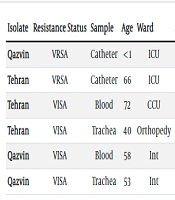
Out of 270 isolates, 264 (97.9%) were sensitive to vancomycin, and 6 (2.1%) were insensitive according to agar dilution and E-test methods. Two (0.74%) isolates had a MIC greater than 256 µg/mL for vancomycin and were identified as vancomycin-resistant S. aureus (VRSA), while 4 (1.4%) isolates were less sensitive to vancomycin and classified as vancomycin-intermediate resistant S. aureus (VISA) (Table 3). The MIC50 for vancomycin in S. aureus isolates was 0.38 μg/mL, and the MIC90 was 4 µg/mL.
| Isolate | Resistance Status | Sample | Age | Ward | Sex | Vancomycin | MICVAN | vanA | MRSA | mecA | mecC | LNZ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Qazvin | VRSA | Catheter | < 1 | ICU | F | Resistant | > 256 | + | Yes | + | - | Sensitive |
| Tehran | VRSA | Catheter | 66 | ICU | M | Resistant | > 256 | + | Yes | + | - | Sensitive |
| Tehran | VISA | Blood | 72 | CCU | F | Intermediate | 4 | - | Yes | + | - | Sensitive |
| Tehran | VISA | Trachea | 40 | Orthopedy | M | Intermediate | 3 | - | Yes | + | - | Sensitive |
| Qazvin | VISA | Blood | 58 | Int | M | Intermediate | 4 | - | No | - | - | Sensitive |
| Qazvin | VISA | Trachea | 53 | Int | F | Intermediate | 4 | - | Yes | + | - | Sensitive |
4.4. Detection of Resistance Genes by PCR and Sequencing
PCR was performed to identify the presence of resistance genes in S. aureus isolates. Two (0.74%) isolates contained the vanA gene (Table 3). However, vanB, vanC, and cfr genes were not detected in any isolates. The mecA and mecC genes were also evaluated, revealing that 144 (53.3%) isolates possessed only the mecA gene. Eight MRSA isolates were negative for the mecA gene (8/152 = 5.3%), and the mecC gene was not detected in any isolates, including the 8 mecA-negative MRSA isolates. The correlation between cefoxitin susceptibility and the presence of mecA was 94.7%.
5. Discussion
The high prevalence of MRSA and the emergence and spread of VRSA and VISA isolates are significant concerns for the health system today. The increasing resistance to methicillin and the ineffectiveness of β-lactam antibiotics in treating MRSA infections have made vancomycin and linezolid the drugs of choice for treating methicillin-resistant staphylococcal infections. According to this research, 152 (56.3%) isolates were identified as MRSA, most of which contained the mecA gene. Two (0.7%) VRSA isolates were detected, both possessing the vanA gene. Additionally, four VISA (1.4%) isolates were identified, none of which contained the vanA gene. All VRSA and VISA strains, except one, were MRSA and encoded the mecA gene. None of the isolates contained the mecC, vanB, or vanC genes. Furthermore, the frequency of isolates resistant to linezolid was very low, and none contained the cfr gene.
The indiscriminate use of antibiotics has led to a growing increase in antibiotic resistance (22). In Iran, the spread of antibiotic resistance in pathogenic bacteria is a significant challenge for the medical community in treating infectious diseases (23). With the global spread of MRSA strains, the management and treatment of MRSA infections have become increasingly important (26). Vancomycin and linezolid are the antibiotics of choice for treating MRSA infections; however, resistance to these antibiotics has also been observed in some cases (24). Since the genes involved in the development of resistance to these two antibiotics can be transmitted horizontally between different strains and species, assessing their frequency in different populations can provide valuable therapeutic insights (25). Therefore, this study investigated resistance to methicillin, vancomycin, and the related genes (vanA, B, C), as well as resistance to linezolid and its related gene (cfr) in S. aureus isolates from patients admitted to Qazvin and Tehran teaching hospitals.
Research in this area indicates varying levels of resistance across different regions globally and within Iran. The first outbreaks of MRSA infections were reported in Europe in 1961 (26). In this study, 152 (56.3%) isolates were identified as MRSA, with 144 (94.7% of MRSA isolates) containing the mecA gene. Eight MRSA (5.3%) isolates did not have mecA or mecC genes, suggesting other resistance mechanisms such as increased expression of PBP2a or specific environmental conditions may be involved, which need further investigation. A total of 56 (20.7%) isolates were identified from the ICU, highlighting the significance of these isolates' prevalence in intensive care units.
According to Shady et al., out of 220 S. aureus isolates, 76 (43.5%) were MRSA (27). Similarly, Ghaderi et al. reported that out of 164 S. aureus isolates, 78 (47.56%) were methicillin-resistant, aligning closely with our study's results (22). Additionally, a study by Khan et al. in Pakistan reported 315 MRSA isolates (65%) out of 485 S. aureus isolates, indicating a high frequency of the mecA gene (23). These differences in results could be attributed to factors such as geographical location, the health settings of various hospital wards, and the infection control practices implemented in these wards. In developing countries, where health policies and care indicators are lower, the prevalence of MRSA strains, particularly in hospitals, is expected to increase.
There is also considerable variability in MRSA prevalence from country to country. Global epidemiology of MRSA shows a prevalence of 67.9% in Europe, 43.2% in the Western Pacific, 31.9% in the Americas, 27.3% in Southeast Asia, 19.1% in Africa, and 17.4% in the Mediterranean. These reports suggest that even though developed countries have higher health and hospital standards, the prevalence of MRSA isolates has been rising, which may be linked to factors such as sample population and collection methods (28).
Currently, 30% to 50% of Staphylococcus isolates are resistant to methicillin, with this resistance spreading globally, including in Iran. Methicillin-resistant strains are impervious to all beta-lactam antibiotics, making vancomycin the primary treatment for infections caused by these strains (24).
The first resistance mechanism involves mutations in the bacterial genome, leading to strains with reduced susceptibility to vancomycin. These strains have been reported worldwide (25). In Iran, Aligholi et al. isolated the first VRSA strain from two patients, both exhibiting a MIC > 256 μg/mL. Only one isolate tested positive for the vanA gene (29). Ziasistani et al. studied 205 S. aureus isolates in Kerman, identifying two VRSA strains in patients with prolonged hospital stays and MIC ≥ 64 μg/mL; both strains carried the vanA gene (30). Dezfulian et al. reported a case of VRSA in a 57-year-old diabetic female, with the isolate showing resistance to multiple antibiotics and a vancomycin MIC of 512 μg/mL. The isolate contained the vanA, R, and S genes (31).
Incidence rates of VRSA strains vary globally. VRSA prevalence is reported to be 16% in Africa, 5% in Asia, 1% in Europe, 4% in North America, and 3% in South America. Asia, particularly Iran and India, has higher reports of VRSA than other continents, underscoring the need for strict antibiotic policies and active monitoring of nosocomial infections. Nigeria (29%) and Saudi Arabia (18%) also report high VRSA prevalence, which is a significant concern (32).
Hadadi et al. investigated 85 S. aureus isolates using the E-test and reported that only one (2%) showed intermediate resistance to vancomycin (VISA) (33). Additionally, a study by Thati et al. in India, which assessed antibiotic resistance in 358 S. aureus samples from an intensive care unit using the agar dilution method, found that 284 (79.6%) isolates were MRSA, 23 (6.5%) were VISA, and seven were VRSA, with six of these VRSA isolates carrying the vanA gene (34). In a study by Aubaid et al. in Iraq, out of 250 clinical specimens of S. aureus, 72 isolates contained the mecA gene, five isolates had the vanA gene, and nine isolates had the vanB gene (35). Asadpour and Ghazanfari investigated 110 S. aureus isolates, finding eight (7.27%) VISA isolates with MICs in the range of 4 - 8 μg/mL, of which only one strain was positive for the vanA gene (6).
In the current study, the minimum inhibitory concentration (MIC) of vancomycin was determined using both the agar dilution method and the E-test. Out of 270 S. aureus isolates, four (1.4%) were identified as VISA and two (0.7%) as VRSA. None of the isolates contained the vanB or vanC genes. The two vancomycin-resistant isolates were sourced from catheters.
It is hypothesized that the mechanism of vancomycin resistance in both VRSA isolates in this study is related to the vanA gene. Consistent with other studies in Iran, this suggests that van gene-dependent vancomycin resistance is on the rise in the country. Although VISA isolates are commonly reported in MRSA strains, they can also exhibit vancomycin resistance in MSSA strains. Therefore, it is essential to screen both MRSA and MSSA isolates to determine the prevalence and frequency of vancomycin resistance.
The MIC results indicate that the four VISA isolates identified in this report have vancomycin concentrations below the threshold recommended in the CLSI agar screen method. This suggests that the agar screen may not be suitable for initial screening to detect resistant strains. In this study, the MIC of vancomycin was confirmed using the agar dilution method and the E-test, revealing four (1.4%) VISA isolates and two (0.7%) VRSA isolates with MICs greater than 256 μg/mL.
The presence of the vanB and vanC genes was negative. Both resistant isolates were obtained from catheter samples. The first sample was identified from a neonatal patient admitted to the ICU of Kosar Hospital in Qazvin, and the second resistant sample was isolated from a 66-year-old male patient admitted to the ICU of Imam Hossein Hospital in Tehran. Both VRSA isolates were resistant to methicillin but sensitive to linezolid. Additionally, two VISA isolates were found in blood and trachea samples from patients hospitalized in Tehran, and two other VISA isolates were obtained from patients admitted to Qazvin hospitals, also from blood and trachea samples. These findings highlight the importance of ICU environments and the use of invasive methods in the prevalence of vancomycin-resistant bacteria.
The results of this study indicated that the prevalence of MRSA strains in ICUs and infectious wards is higher than in other wards. This can be attributed to the long-term hospitalization of patients in these wards, the overuse of antibiotics, and the use of invasive methods, leading to the emergence of resistant strains. Bijari et al. showed that the high frequency of MRSA in ICUs, about 75%, is a serious concern for hospitals (36). Honda et al. reported that MRSA was more common in ICUs, accounting for 66.4% of ICU-acquired S. aureus infections (37). In the Chi et al. study, S. aureus was identified as the most common pathogen among ICU patients with Ventilator Associated Pneumonia (VAP) (38). These findings are consistent with the results of the present study.
This study has several limitations. While we identified the mechanism of resistance to linezolid through the cfr gene, we were unable to investigate other mechanisms contributing to resistance to this antibiotic. Additionally, van genes are located on specific transposons (Tn-1546 and Tn-1549 transposons) found on plasmid PLw1043. These mobile genetic elements play a crucial role in transferring resistance genes between S. aureus isolates, but we could not identify these elements in this study, representing another limitation.
Staphylococcus aureus with intermediate resistance to vancomycin (VISA) suggests that factors other than the presence of van genes can contribute to this resistance, such as mutations in genes like rpoB, vrasR, walkR, and the yvqF/vraSR genetic system. Many of these mutations are directly or indirectly involved in the metabolism and biosynthesis of the staphylococcal cell wall, leading to chromosomal resistance to vancomycin, which was not investigated in our study as it focused on resistance related to mobile genetic elements. Future studies should investigate these factors.
5.1. Conclusions
The results of this study indicate that the prevalence of MRSA strains in ICUs and infectious wards is higher than in other hospital wards. We found that 4 (1.4%) isolates exhibited intermediate resistance to vancomycin (VISA), and 2 (0.7%) isolates were resistant to vancomycin (VRSA), which is concerning. Given that the resistance gene is located on transposable elements (transposons), the potential for transmission to other bacteria is high. Although the number of MRSA strains is increasing, our study demonstrates that vancomycin and linezolid remain effective drugs of choice due to the low resistance of clinical strains to these antibiotics. Emphasizing follow-up programs to control and restrict the spread of MRSA strains is vital. Given the high clinical significance of infections caused by S. aureus, it is essential to inform physicians and medical staff and to develop meticulous methods for identifying and controlling such infections within medical systems.