1. Background
Despite the availability of an effective vaccine and potent antiviral treatments, hepatitis B virus (HBV) infection is still a major global health problem (1, 2). By 2015, approximately 2 billion individuals had been exposed to HBV, with nearly 257 million estimated cases of chronic hepatitis B infection (3). Hepatitis B Virus and related conditions are also thought to be responsible for 500,000 to 700,000 deaths, annually (2). It is highly endemic in sub-Saharan Africa and East Asia, where approximately 5% to 10% of the adult population is chronically infected (4). In Kenya, the seroprevalence of HBsAg positivity is about 5% to 14% and the prevalence of HBV exposure is 20% to 70% (5, 6). For this reason, treatment and follow-up of chronic HBV infection is crucial.
Furthermore, HBV replication or detectable serum HBV DNA is closely related to the progression of liver disease, liver function decompensation, and the occurrence of cirrhosis and liver cancer in patients with chronic hepatitis B (CHB) infection (7).
Detection of serological markers is the backbone for the diagnosis of HBV infection, and quantification of HBV surface Antigen (HBsAg) in serum has been identified as the most reliable marker of HBV carriage (8). Hepatitis B Virus e antigen (HBeAg) is generally used as a secondary marker to indicate high levels of virus in the blood (8). The quantitative levels of HBV DNA, alanine aminotransferase (ALT), and histological outcomes constitute a major component in the management and treatment of HBV (9). The monitoring of HBV DNA in serum is as important as serological markers in predicting the clinical outcome of infection. Molecular diagnostic methods have been used to quantify the levels of HBV DNA in serum as a marker of viral replicative activity (9). The detection and quantification of HBV DNA is reported to have prognostic value for the outcomes of acute and chronic HBV infections (6). However, HBV DNA quantification assays are molecular-based and expensive (9). Given the high prevalence of HBV, particularly in developing countries, cheaper tests that can be used to monitor viral replication and response to antiviral therapy would make CHB management more practical.
Focus has shifted to the quantification of HBsAg due to its association with the level of covalently closed circular (ccc) DNA showing viral replication inside the nuclei of hepatocytes (10). Positive correlations have been documented between HBsAg and serum HBV DNA levels in patients with CHB on treatment (11, 12). Baseline HBsAg levels have been shown to predict both HBsAg seroclearance (13) and a surrogate marker for monitoring HBV treatment response (14).
Although HBV treatment by HBV DNA monitoring is done in selected high cost hospitals and laboratories in Kenya, this study investigated whether, using serum HBsAg concentration as a marker of HBV replication level in patients with hepatitis B, could correlate with HBV DNA levels in CHB in Nairobi, Kenya.
2. Methods
2.1. Study Design and Setting
This cross-sectional study was conducted on patients with HBV referred to the Pathologist Lancet Laboratory in Nairobi, Kenya. Using the Lemeshow et al. (15) formula for estimating population proportion with specified absolute precision, setting α at 0.05, and correlation rates of 34% (16) for HBsAg and HBV DNA levels, a total of 172 HBV patients were recruited to achieve 0.95 power and to obtain a total of 344 matched HBsAg and HBV DNA level tests. Consenting adult patients were enrolled in the study. A standardized questionnaire was used to collect demographic data while 5 mL of blood was drawn from each patient for one-time HBsAg and HBV DNA quantification.
2.2. Ethical Approval and Consent to Participate
This study was conducted according to the declaration of Helsinki and international conference on harmonization guideline on good clinical practice. The protocol and informed consent form were reviewed and approved by the Kenyatta University Scientific Steering Committee and the Ethical Review Committee prior to commencement of field activities (PKU/304/E280 on 25th February, 2015). Written informed consent was obtained from each participant.
2.3. Laboratory Analysis
Quantification of HBsAg was performed by an automated chemiluminescent micro-particle immunoassay using the Roche Cobas e 411 analyzer with Elecsys HBsAg II Quant reagent kits (Roche Diagnostics, CA, USA), according to the manufacturer’s instructions. Quantification of HBeAg was conducted by an automated chemiluminescent micro-particle immunoassay using the Architect platform (Abbott Diagnostic, Chicago, IL, USA), according to the manufacturer’s instructions. Serum HBV DNA levels (copies/mL) were measured by polymerase chain reaction (PCR) using CobasAmpliPrep/CobasTaqMan HBV Test, version 2.0 (Roche Molecular Systems, CA, USA), according to the manufacturer’s instructions.
2.4. Statistical Analyses
Quantitative levels of HBsAg, HBeAg, and HBV DNA were expressed as the mean ± standard deviation (SD). A comparison of HBsAg and HBV DNA between HBeAg positive and negative patients was performed using the Mann-Whitney U test. Spearman correlation coefficient was used to correlate serum levels of HBsAg and HBV DNA levels. Data analysis was done using the SPSS software for Windows, v.21 where P ≤ 0.05 was considered significant.
3. Results
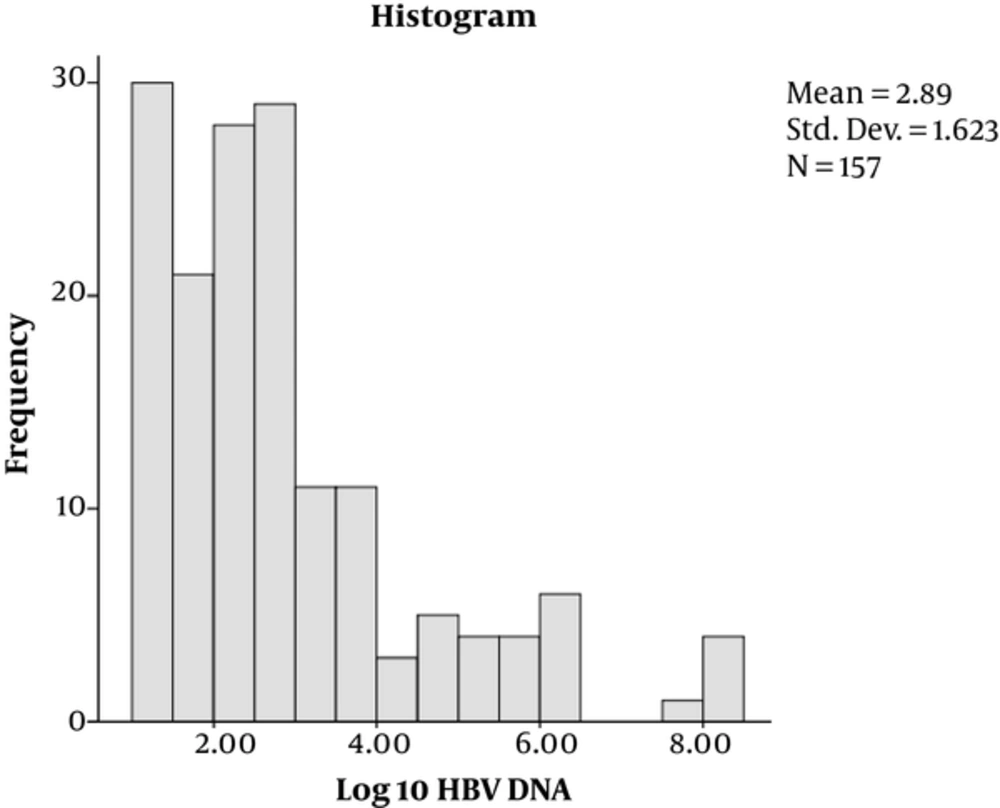
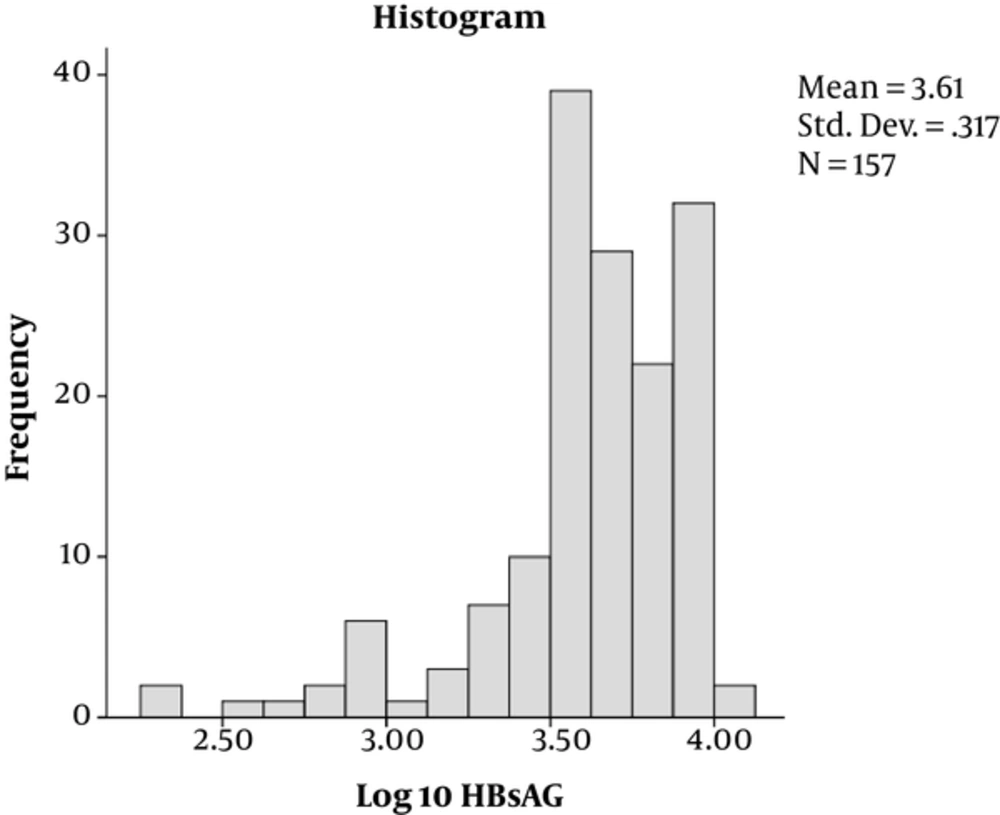
Out of 173 enrolled participants, the majority, 112 (64.7%), were male while 61 (35.3%) were female. Their mean age was 37.17 years with majority of the patients, 109 (68.8%), aged below 40 years. Of the 173 patients, 142 (82.1%) were HBeAg-negative while 31/173 (17.9%) were positive (Table 1). By Kolmogorov-Smimov test, the distribution of HBsAg titers and HBV DNA levels were irregular (Figures 1 and 2, respectively).
| Variable | Patients | Mean ± SD | P Value |
|---|---|---|---|
| Age, y | 37.2 ± 11.1 | 0.001a | |
| 20 - 30 | 53 (30.6) | ||
| 31 - 40 | 66 (38.2) | ||
| 41 - 50 | 26 (15) | ||
| ≥ 51 | 28 (16.2) | ||
| Gender | 0.001a | ||
| Male | 112 (64.7) | ||
| Female | 61 (35.3) | ||
| HBeAg-positive CHB | 31 (17.9) | 0.001a | |
| HBeAg-negativ CHB | 142 (28.1) | 0.001a | |
| Log 10-transformed overall HBsAg titer, IU/mL | 3.58 ± 0.39 | ||
| Log 10-transformed HBV-DNA, IU/mL | 2.89 ± 1.62 | 0.032b | |
| Log 10-transformed HBsAg titer, IU/mL in HBeAg-positive CHB | 3.67 ± 0.37 | ||
| Log 10-transformed HBsAg titer, IU/mL in HBeAg-negative CHB | 3.56 ± 0.41 | ||
| Log 10-transformed HBV-DNA, IU/mL in HBeAg- positive CHB | 5.17 ± 1.58 | 0.001b | |
| Log 10-transformed HBV-DNA, IU/mL in HBeAg- negative CHB | 2.07 ± 1.27 | 0.001b |
Abbreviations: CHB, chronic hepatitis B; HBV-DNA, hepatitis B virus-deoxyribonucleic acid; SD, standard deviation.
aExact test.
bMann-Whitney.
The overall mean (Log10) of HBsAg (SD ±) titer was 3.58 ± 0.39 IU/mL and an overall mean (Log10) of HBV-DNA (SD ±) of 2.89 ± 1.62 IU/mL (Table 1).
The mean (Log10) of HBsAg (SD ±) titer in CHB, in those who were HBeAg positive and negative, was 3.67 ± 0.37 IU/mL and 3.56 ± 0.41 IU/mL, respectively (Table 1).
The mean (Log10) of HBV-DNA (SD ±) in HBeAg-positive patients and HBeAg-negative patients were 5.17 ± 1.58 and 2.07 ± 1.27 IU/mL, respectively. By Mann-Whitney test, HBV DNA levels differed significantly between HBeAg positive and negative patients (P = 0.001), as did HBsAg titer (P = 0.032) (Table 1).
Overall, a weak but significant correlation was found between serum quantitative HBsAg and HBV DNA (r = 0.171; P = 0.024). Quantitative HBsAg was not significantly correlated with HBV-DNA among the HBeAg positive patients (r = 0.309; P = 0.093). Similarly, no significant correlation was found among the HBeAg negative patients (r = 0.065; P = 0.443) (Table 2).
| Variable | HBsAg, IU/mL | HBV-DNA, IU/mL | r | P Valuea |
|---|---|---|---|---|
| All subjects (n = 173) | 3.58 ± 0.39 | 2.89 ± 1.62 | 0.171 | 0.024 |
| HBeAg positive (n = 31) | 3.67 ± 0.37 | 5.17 ± 1.58 | 0.311 | 0.089 |
| HBeAg negative (n = 142) | 3.56 ± 0.41 | 2.07 ± 1.27 | 0.065 | 0.443 |
Abbreviation: SD, Standard deviation.
aSpearman’s correlation test; significant at the 0.05 level (2-tailed).
4. Discussion
This study attempted to determine the correlation between the HBsAg levels and those of HBV DNA levels among HBV patients attending one of the largest private referral laboratories in Nairobi, Kenya. HBsAg is a classical marker of infection with hepatitis B virus, and serological assays to detect HBsAg have guided its diagnosis. This study hypothesized that quantification of HBsAg could be used to manage and monitor HBV patients. Upon HBV infection, closed circular HBV DNA genome forms within the hepatocytes nuclei, which becomes enveloped and secreted into the blood (13). Furthermore, HBV DNA, is the template for gene transcription and replication, correlating robustly with levels of total intracellular HBV DNA, serum HBV DNA, and HBsAg (14). Thus, HBV DNA level is the most important and most direct etiological evidence for HBV (17). However, in developing countries endemic for HBV (18), robust, cheaper, and easy-to-perform markers are essential in the management and monitoring of HBV patients (19). Hepatitis B viral load measurement is rarely accessible and where available often done once, which may not reflect real viral activity (19). Furthermore, usefulness of viral load in disease monitoring, and medication in patients with undetectable HBV DNA levels is still missing (20). Quantification of HBsAg indirectly reflects the number of infected hepatocytes (21) and is known to change over the natural course of chronic HBV infection. Hepatitis B surface Antigen, a membrane protein of the HBV envelope, has been proposed as a marker for monitoring HBV infected patients (22). During antiviral therapy, HBsAG quantification could be used to differentiate true inactive carriers from patients in remission, who are likely to progress to cirrhosis (23). Tsai et al. (24) showed that HBV DNA positivity could be easily detected in patients with higher levels of serum HBsAg than those with lower levels.
The current study found a weak but significant overall correlation between HBsAg and HBV DNA levels (r = 0.171, P < 0.001). However, there was no significant correlation between HBsAg level and HBV DNA level among HBeAg positive and negative patients (r = 0.309, P = 0.093; r = 0.065, P = 0.443, respectively). This study mirrors the report by Ganji et al. (9) who found no significant correlation between HBsAg and HBV DNA levels among CHB (r = 0.53; P = 0.606), including HBeAg positive and negative patients (r = -0.57; P = 0.053 and r = 0.057; P = 0.605 respectively). Similar results were also reported by Mahdavi et al. (25) in the overall population (r = 0.231, P = 0.656). Demiroren et al. (12) also showed an overall negative non-significant correlation (r = -0.113, P = 0.413). On the contrary, several reports have showed a positive correlation between HBsAg and HBV DNA level in the entire cohort, HBeAg-positive and -negative groups. Togo et al. (11) reported an overall positive correlation (r = 0.586, P < 0.001). Kim et al. (26) reported a slightly higher significant correlation (r = 0.693, P < 0.001), and this was also observed in HBeAg-positive and -negative patients. Teriaky and Al-Judaibi (27) showed a significant positive correlation in the entire cohort, HBeAg-positive and -negative groups (r = 0.402, P < 0.001; r = 0.383, P < 0.05; r = 0.309, P < 0.01, respectively). Zhu and Zhang (20) reported an overall positive correlation (r = 0.657, P < 0.05) while Jaroszewicz et al. (28) showed even a stronger significant correlation between HBsAg and HBV-DNA in the overall population (r = 0.79, P < 0.01).
The variations in the correlation between HBsAg and HBV-DNA levels was attributed to differences in disease stage, HBeAg status, HBV genotypes involved, and types/phases of treatment (16, 25) and should always be considered in HBV DNA and HBsAG level correlation studies. Even though most of these factors were not established in the current study, similar to the report by Ganji et al. (9), this study highlights a significant insight into differences in HBsAg levels between HBeAg positive and negative patients, which appear to be affected by HBeAg status. Hepatitis B Virus DNA and HBsAg levels were higher in HBeAg positive patients.
Although not a focus of this study, several biochemical parameters including aspartate aminotransferase (AST), alanine aminotransferase (ALT) albumin, total bilirubin, gamma-glutamyl transferase (GGT), alpha-fetoprotein (AFP) and platelet count have used for the assessment of HBV infection and treatment outcomes (29). Variations in the correlation between HBV DNA levels and biochemical parameters have been reported; some showing positive correlations (12) while others showing contrary outcomes (9). This should complement future HBsAg and HBV-DNA correlation studies in Kenya.
4.1. Limitations and Conclusions
This study had several limitations including (i) the small sample size of the HBeAg – positive patients, which may have limited the statistical significance obtained, (ii) the cross sectional nature; it was not possible to measure serially the HBsAg and HBV DNA levels, which have been shown to fluctuate especially among chronic HBeAg-negative HBV-infected patients, (iii) HBV DNA and no transcriptionally active covalently closed circular (ccc) HBV DNA were measured, which has been shown to correlate more appropriately with HBsAg (30). The stage of disease, HBV genotypes and types/phases of treatment were not determined and we cannot rule out their role in the overall weak or the lack of correlation in the levels of HBV DNA and HBsAG in the entire patient population or either HBeAg-positive or HBeAg-negative patients, respectively.
Given these limitation, this study provided an insight on to differences in HBV DNA and HBsAg levels between HBeAg-positive and -negative patients, which appear to be affected by HBeAg status. Serum quantitation of HBsAg may not replace serum HBV DNA levels among CHB patients in Nairobi Kenya.