1. Background
Staphylococcus aureus, as one of the most important nosocomial pathogens, is known to be responsible for a diverse array of infections. Several studies have recently focused on understanding the importance of expressed S. aureus genes, which are involved both in virulence and antibiotic resistance (1). The pathogenesis of this bacterium is linked to the expression of various virulence determinants. An important virulence factor identified in this microorganism is Panton-Valentine leukocidin (PVL) (1, 2). These strains are particularly important because they are primarily associated with wound and life-treating infections. The role of PVL in the pathogenesis of S. aureus is clear; however, whether the toxin affects disease severity, clinical presentation, and outcome is a matter of debate (1-3).
Most clinical and epidemiological studies have focused on PVL as a potential marker for community-associated methicillin-resistant S. aureus (MRSA) infections. Some studies have recently attempted to address that PVL genes are also related to healthcare-associated methicillin-resistant S. aureus (HA-MRSA) strains that indicate a change in the epidemiology of PVL-positive S. aureus isolates (4).
Following efforts made to map out the prevalence and spread of different PVL-positive S. aureus types in different parts of the world, Shahini Shams Abadi et al., in a systematic review and meta-analysis, indicated the prevalence of PVL among S. aureus isolates obtained from cutaneous infections within the range of 7.4 - 55.6% (3). However, there have also been reports of a concurrent worldwide increase in the prevalence of PVL among S. aureus isolates in different parts of the world (3, 5-9). Although the prevalence and distribution of hospital-associated (HA)-S. aureus carrying pvl genes have been well established, but there is a shortage of data on the characteristics of the PVL-positive S. aureus lineages in Iran.
2. Objectives
Given the high frequency of PVL in HA-S. aureus strains and their genetic diversity, this study set out to establish the frequency, antibiotic resistance, and genetic characteristics of PVL-positive HA-S. aureus strains isolated from clinical samples.
3. Methods
3.1. Sample Collection and Staphylococcus aureus Isolation
The 65 S. aureus carrying pvl genes used in the present study were obtained from 345 S. aureus isolated from five hospitals during the period of 2 years from July 2020 to June 2022. To initially identify the isolates, bacteriological and biochemical techniques were recruited (10). For definite identification, polymerase chain reaction (PCR) for the nucA gene was performed, and for the next step, the presence of the pvl genes was assessed using PCR assay (11).
3.2. Determination of Isolates’ Susceptibility
The Kirby-Bauer disk diffusion method was performed under the Clinical and Laboratory Standards Institute (CLSI) criteria for the susceptibility evaluation of isolates against penicillin, tetracycline, rifampin, clindamycin, quinupristin-dalfopristin, erythromycin, ciprofloxacin, nitrofurantoin, linezolid, gentamicin, and trimethoprim-sulfamethoxazole (12). Susceptibility to vancomycin and mupirocin (i.e., low-level mupirocin resistant (LLMUPR) and high-level mupirocin resistance (HLMUPR)) was determined by the broth microdilution test according to the CLSI guidelines. The D-zone test was performed to detect inducible clindamycin resistance S. aureus isolates. Reference strains of S. aureus ATCC 25923, ATCC 43300, and ATCC 29213 were used to control the experiment. Multidrug-resistant (MDR) isolates cover resistance to three or more unique antibiotic classes.
3.3. Extraction of Genomic DNA
DNA was extracted from the isolates identified as PVL-positive S. aureus using the phenol-chloroform method, along with adding lysostaphin (Sigma-Aldrich Co., USA) at a final concentration of 30 µg/mL for the cell wall lysis as previously described. The purity of DNA was monitored by a spectrophotometer (10).
3.4. MRSA Screening
The in vitro evolution of methicillin resistance was performed with a cefoxitin disc (30 µg) on Mueller-Hinton agar plates under the CLSI guideline (12). The mecA-mediated resistance was detected using PCR, as previously described (10).
3.5. Genotypic Characterization
3.5.1. Staphylococcal Cassette Chromosome mec Typing
A multiplex PCR was recruited for identifying staphylococcal cassette chromosome mec (SCCmec) types based on the oligonucleotide sequences and conditions described by Boye et al. (13). Obtained banding patterns were analyzed by comparing them to the banding patterns of reference stains as follow:
ATCC 10442 (SCCmec type I), N315 (SCCmec type II), 85/2082 (SCCmec type III), MW2 (SCCmec type IVa), and WIS (SCCmec type V) as reference strains
3.5.2. Detection of agr Alleles
To amplify the hypervariable domain of the agr locus and subsequently typing of isolates, multiplex PCR was performed based on the Gilot et al. method (14). The banding patterns of each isolate were compared to agr reference strains as follows:
The agr group-I strains with a 441-bp fragment, agr group-II strains with a 575-bp fragment, agr group-III strains with a 323-bp fragment, and agr group-IV strains with a 659-bp fragment
4. Results
4.1. Participants, Isolation, and Screening of PVL-Positive Strains
In the current survey, 345 S. aureus isolates were collected from five hospitals (A-E) during the study period, 18.8% (n = 65) of which were PVL-positive isolates. The studied isolates comprised mixed specimen types; the majority of PVL-positive isolates were obtained from a wound, representing 43.1% (28/65); however, other sources included blood (23.1%; 15/65), purulent discharge (15.4%; 10/65), urine (7.7%; 5/65), sputum (6.1%; 4/65), and body fluids (4.6%; 3/65). The majority of PVL-positive S. aureus strains were isolated from hospital C (41.5%, 27/65), followed by hospital B (23.1%; 15/65), hospital A (15.4%; 10/65), hospital E (12.3%; 8/65), and hospital D (7.7%; 5/65). The distribution of strains in different wards included 18 (27.7%), 15 (23.1%), 13 (20%), 12 (18.5%), and 7 (10.7%) isolates from the intensive care unit, surgery, internal, infectious, and oncology wards, respectively.
4.2. Antimicrobial Resistance Profiles of PVL-Positive Isolates
As observed in Table 1, all the isolates were susceptible to linezolid. None of the isolates under study was susceptible to all of the antibiotics. The resistance rate of penicillin (PEN) was the highest (93.8%), followed by erythromycin (ERY) (84.6%), tetracycline (TET) (72.3%), ciprofloxacin (CIP) (70.8%), rifampin (RIF) (64.6%), gentamicin (GEN) (63.1%), clindamycin (CLI) (49.2%), nitrofurantoin (NIT) (44.6%), trimethoprim-sulfamethoxazole (SXT) (35.4%), mupirocin (MUP) (32.3%), quinupristin-dalfopristin (SYN) (27.7%), and vancomycin (VAN) (12.3%). Most of the PVL-positive isolates were confirmed as MRSA (66.2%); nevertheless, methicillin-susceptible S. aureus (MSSA) was detected in 33.8% of the isolates. All S. aureus strains isolated from urine (n = 5) and sputum (n = 4) were MSSA. The MRSA isolates showed increased resistance rates over MSSA isolates to examined antibiotics. Regarding MSSA isolates, higher resistance rates were recorded against PEN (30.8%), RIF (24.6%), and ERY (23.1%). None of the MSSA isolates was resistant to MUP and VAN. Higher resistance rates among MRSA isolates belonged to PEN (63.1%; 41/65), TET (61.5%), and ERY (61.5%). In total, 84.6% of the isolates were found to be MDR, 40 MRSA and 15 MSSA isolates of which verified to be MDR accounted for 61.5% and 23.1%, respectively.
| Simultaneous Resistance to Antibiotics and Resistance Profile | Resistance Pattern | Type of Samples (n; % Indicated When Not 100%) | MRSA/MSSA (n; % Indicated When Not 100%) | Number of Isolates (%) |
|---|---|---|---|---|
| Eleven | ||||
| A | PEN, TET, CIP, RIF, NIT, GEN, ERY, CLI, SYN, SXT, MUP | W (6; 66.7), B (3; 33.3) | MRSA | 9 (13.8) |
| B | PEN, TET, RIF, NIT, GEN, ERY, CLI, SYN, SXT, MUP, VAN | W (3) | MRSA | 3 (4.6) |
| Seven | ||||
| C | PEN, TET, CIP, RIF, GEN, ERY, CLI | W (3; 30), PD (4; 40), U (2; 20), BF (1; 10) | MRSA (5; 50), MSSA (5; 50) | 10 (15.4) |
| D | PEN, TET, CIP, GEN, ERY, SXT, VAN | W (1; 20), BF (1; 20), B (3; 60) | MRSA | 5 (7.7) |
| E | PEN, TET, CIP, RIF, NIT, GEN, ERY | W (9; 69.2), B (3; 23.1), U (1; 7.7) | MRSA (6; 46.2), MSSA (7; 30.8) | 13 (20) |
| Six | ||||
| F | PEN, TET, CIP, ERY, CLI, MUP | W (2; 28.6), PD (2; 28.6), B (2; 28.6), BF (1; 14.2) | MRSA | 7 (10.8) |
| G | PEN, ERY, CLI, SYN, SXT, NIT | W (1; 33.3), B (2; 66.7) | MRSA | 3 (4.6) |
| Four | ||||
| H | PEN, ERY, SYN, SXT | S (2; 66.7), U (1; 33.3) | MSSA | 3 (4.6) |
| Three | ||||
| I | CIP, ERY, MUP | PD (2) | MRSA | 2 (3.1) |
| J | PEN, NIT, GEN | U (1) | MSSA | 1 (1.5) |
| Two | ||||
| K | PEN, RIF | W (3; 42.8), PD (2; 28.6), B (2; 28.6) | MRSA (3; 42.9), MSSA (4; 57.1) | 7 (10.8) |
| Without | ||||
| L | - | S (2) | MSSA | 2 (3.1) |
Resistance Combinations of Panton-Valentine Leukocidin-Positive Strains and Their Distribution Among Clinical Samples
As shown in Table 1, 11 resistance profiles were detected, wherein PEN, TET, CIP, RIF, NIT, GEN, and ERY (20%; 13/65), PEN, TET, CIP, RIF, GEN, ERY, and CLI (15.4%; 10/65), and PEN, TET, CIP, RIF, NIT, GEN, ERY, CLI, SYN, SXT, and MUP (13.8%; 9/65) were the top 3 frequently detected profiles. The frequency of inducible and constitutive CLI resistance among PVL-positive MRSA strains (12.3% and 26.1%) was higher than PVL-positive MSSA strains (7.7% and 15.4%). In the present study, 32.3% of the isolates were confirmed as MUP resistant, and all were MRSA, 9 (42.9%) and 12 (57.1%) isolates of which exhibited HLMUPR and LLMUPR phenotypes.
4.3. Molecular Characterization
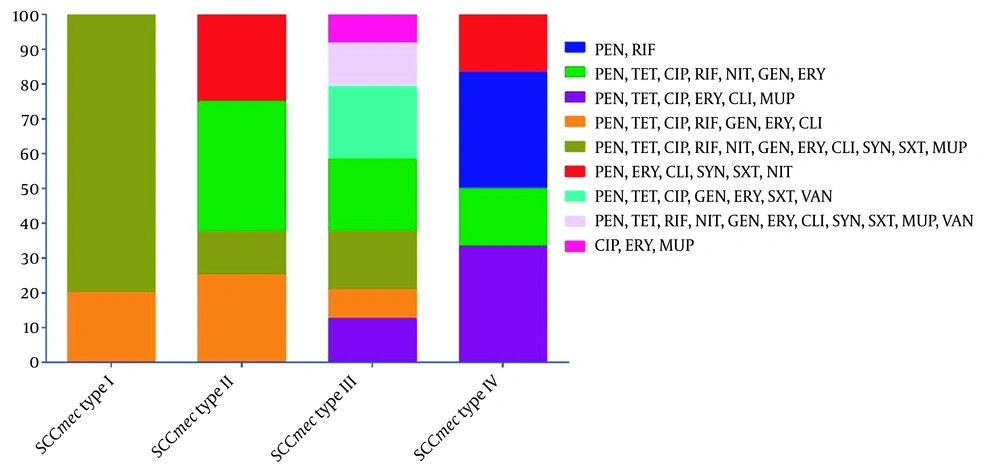
The SCCmec typing results showed that the most prevalent SCCmec type was III, representing 55.8% (24/43); nonetheless, 8 (18.6%), 6 (14%), and 5 (11.6%) isolates expressed SCCmec types II, IV, and I, respectively. Staphylococcal cassette chromosome mec type I was recovered from blood (40%; 2/5), wound (40%; 2/5), and body fluid (20%; 1/5). Staphylococcal cassette chromosome mec types II were recovered from blood (37.5%; 3/8), wound (12.5%; 1/8), and purulent discharge (50%; 4/8). Staphylococcal cassette chromosome mec types III were recovered from blood (33.3%; 8/24), wound (54.2%; 13/24), body fluid (4.2%; 1/24), and purulent discharge (8.3%; 2/24). Staphylococcal cassette chromosome mec types IV were recovered from the wound (83.3%; 5/6) and body fluid (16.7%; 1/6) samples. All 8 vancomycin-resistant MRSA isolates belonged to SCCmec III, 5 (62.5%) and 3 (37.5%) isolates of which had constitutive and inducible clindamycin resistance phenotypes, respectively. All HLMUPR MRSA isolates belonged to SCCmec III and recovered from the wound samples. The LLMUPR accounted for 6.2% (4/65), 1.5% (1/65), 4.6% (3/65), and 3.1% (2/65) of SCCmec types I, II, III, and IV, respectively. Figure 1 depicts a summary of the distribution of resistance profiles among different SCCmec types.
Distribution of resistance profiles in different staphylococcal cassette chromosome mec (SCCmec) types. Abbreviations: PEN, penicillin; RIF, rifampin; TET, tetracycline; CIP, ciprofloxacin; NIT, nitrofurantoin; GEN, gentamicin; ERY, erythromycin; CLI, clindamycin; MUP, mupirocin; SYN, quinupristin-dalfopristin; SXT, trimethoprim-sulfamethoxazole; VAN, vancomycin.
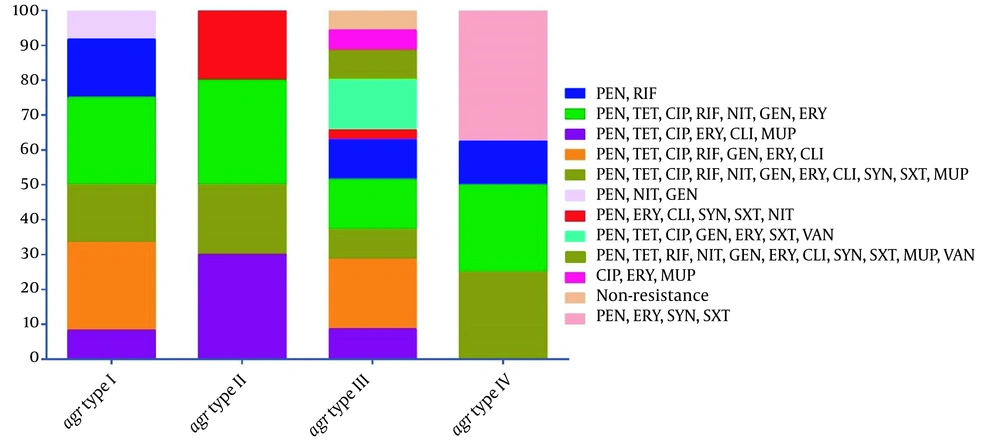
According to agr typing technique, 35 (53.8%), 12 (18.5%), 10 (15.4%), and 8 (12.3%) isolates harbored agr types III, I, II, and IV, respectively. The obtained results revealed that predominantly PVL-positive MRSA strains expressed agr type III (55.8%; 24/43), followed by types I and II (18.6%; every 8 isolates/43) and IV (7%; 3/43); however, PVL-positive MSSA strains predominantly harbored agr types III (50%; 11/22), IV (22.7%; 5/22), I (18.2%; 4/22), and II (9.1%; 2/22). Figure 2 depicts a summary of the distribution of resistance profiles among different agr types. The agr type I was not detected in any PVL-positive S. aureus strains recovered from blood and sputum samples. Most agr type III isolates were isolated from the wound (34.3%; 12/35) and blood (34.3%; 12/35) samples.
Distribution of resistance profiles in different agr types. Abbreviations: PEN, penicillin; RIF, rifampin; TET, tetracycline; CIP, ciprofloxacin; NIT, nitrofurantoin; GEN, gentamicin; ERY, erythromycin; CLI, clindamycin; MUP, mupirocin; SYN, quinupristin-dalfopristin; SXT, trimethoprim-sulfamethoxazole; VAN, vancomycin.
5. Discussion
This survey demonstrated several striking results, including a relatively high frequency of PVL-positive S. aureus strains and a high diversity of their types. This study also showed a high frequency of PVL-positive S. aureus with simultaneous resistance to antibacterial agents. It manifests as a prominent threat in clinical settings. Another finding of the present study was the high resistance prevalence to vancomycin among S. aureus isolates.
The frequency of PVL among S. aureus in this study was 18.8%, a higher prevalence than that observed (4.9%) in S. aureus-related infections in the United Kingdom (7). However, various percentages of PVL-carrying S. aureus were reported by several studies worldwide (5-8). A high prevalence of PVL-carrying S. aureus strains has also been reported in Tunisia (79%) (8), Uganda (49.3%) (5), and Saudi Arabia (30%) (6). The findings of the current survey indicated that the prevalence of PVL-carrying S. aureus in Iran is quite similar to the previously reported rates of 21.4% and 19.5% in 2016 and 2017 in Iranian hospitals, respectively (11). The results pointed to a conclusion that despite the low prevalence of PVL positive, special attention for the laboratory routine detection of these isolates is needed because the mobility of the pvl gene across MRSA isolates might increase the morbidity of nosocomial infections caused by HA-MRSA (15-17).
Mupirocin is used to control the dissemination of S. aureus isolates in communities and healthcare settings and the occurrence of severe infections (18-21). In the present survey, the data showed a high prevalence rate of mupirocin-resistant MRSA isolates (32.3%). This finding is supported by the observations of Chamon et al. from Brazil (33%) (17) and Goudarzi et al. from Iran (30.5%) (11). In a recent meta-analysis study by Dadashi et al., various prevalence rates of mupirocin-resistant MRSA isolates were reported in different geographic areas (e.g., less than 1.0% in France, India, Iran, and Australia or more than 50% in India, the USA, and Egypt) (19). The reason for the high mupirocin resistance rate is not well-understood but can be attributed to the ward, sample type, geographic or socioeconomic factors, and policies in the use of mupirocin in hospitals.
Furthermore, this study reported a high prevalence of HLMUPR and LLMUPR at 42.9% and 57.1% among MRSA isolates, respectively. A recent study in Egypt reported a high prevalence of HLMUPR and LLMUPR, with a proportion of 61.5% and 38.5% (22). In the present survey, the prevalence rates of high and low levels of resistance to mupirocin in MRSA isolates were reported as 13.5% and 18.5%, respectively. The reported rate of HLMUPR-MRSA in this study was higher than in France (0.8%) (20), Canada (4.3%) (18), and China (7%) (21). Overall, the reason for this high mupirocin resistance rate is not well-understood; however, it appears to be related to a shortage in the implementation of antibiotic stewardship programs, incorrect policies unrestricted, and widespread use of this antibiotic. Therefore, before mupirocin therapy, it is essential to determine the susceptibility of isolates to mupirocin.
Methicillin-resistant S. aureus is important; nevertheless, the emergence of vancomycin-resistant S. aureus (VRSA) represents an additional challenge and concern for controlling staphylococcal infections (23). It is worth noting that VRSA was found in 12.3% of PVL-positive isolates. The results of a meta-analysis performed in 2020 depicted an upward trend in VRSA and vancomycin-intermediate S. aureus (VISA) worldwide. Shariati et al. demonstrated that VISA strains (1.7%) had a higher global rate than VISA strains (1.5%). Likewise, an increasing trend of 2 and 3.6-fold of VRSA and VISA after 2010, compared to before that, was noted. However, Asian countries, especially Iran and India, included the highest rates of VRSA incidence (67%) (23). This relatively high prevalence of VRSA in the two aforementioned countries, compared to American/European countries, can be due to the unrestricted and unscheduled administration of antimicrobials, geographic area, level of hygiene, poor health policies, and diverse attitudes toward antimicrobial protocols.
The present study’s observations about SCCmec types are in line with those of other studies that confirmed the relationship of SCCmec types I, II, and III with HA-S. aureus infections; nevertheless, IV and V are prominent types in community-associated (CA)-S. aureus infections (24, 25). Staphylococcal cassette chromosome mec typing illustrated a dominance for type III at 55.8%, which is similar to the results of several studies indicating PVL-positive HA-MRSA strains carrying SCCmec types II and III. This finding indicated that PVL-positive strains with SCCmec type III spread in different regions of Iran. Contrary to the finding of a study by Chamon et al. (17) from Brazil which reported a predominance of one SCCmec IV in PVL-positive S. aureus strains, representing 62% of the isolates, the current study’s observations indicated SCCmec type IV at a low level (14%). This emergence has been reported regionally to higher levels in reports of the nearby countries, with dominance in SCCmec type IV ranging from 19% to 90% (26).
Although numerous studies have characterized agr types of HA- and CA-S. aureus isolates from the community and hospitals; narrow studies have focused on the features description of agr types among PVL-positive S. aureus strains (8, 10). The results of agr typing performed for strains also showed that agr type I was the second predominant genotype in PVL-positive strains. In a study in Thailand on 92 S. aureus strains, this type was more frequent in the tested isolates (27). Another study conducted by Javdan et al. on 150 S. aureus isolates showed that agr type I was predominant (54.7%), followed by type II (24.7%), type IV (14%), and type III (6.6%) (28). The present study observed relatively low infection rates of 15.4% and 12.3% of agr types II and IV, respectively. Similar rates were reported by Ghasemian et al. (29). It could be speculated that agr type I can have a crucial task in the regulation of staphylococcal toxins, especially PVL. The high prevalence of agr type III among PVL-positive isolates promoted us to understand its virulence.
5.1. Conclusions
Overall, this study indicated a diversity of agr and SCCmec types, easily transferred “from and to” hospitals. The widespread dissemination of MDR PVL-positive S. aureus strains was a wake-up call for researchers. Resistance to vancomycin in this study emphasized that using this and other antibiotics, especially mupirocin and clindamycin, and resistance to them should be carefully monitored.