1. Background
Urinary tract infections (UTIs) are among the most common pathological problems, with an annual incidence of 150 million people worldwide (1). They are considered the second most common infection affecting humans, significantly increasing morbidity and mortality (2). Although UTIs occur in both inpatient and outpatient settings, affecting both genders and all age groups (3), women are considered more susceptible to this infection due to the inherited anatomy of the female genitourinary tract (4). About 50% of women and 12% of men suffer from UTIs at least once during their lifetime, and approximately 25% of those affected women will experience a UTI recurrence within 6 - 12 months after the original infection (5).
The UPEC strains are considered the major etiological agents for both complicated and uncomplicated UTIs, accounting for 65% and 75% of cases, respectively (6). In addition, they are responsible for up to 90% of community-acquired UTIs and 50% of nosocomial UTIs (7).
The successful colonization of uropathogenic Escherichia coli (UPEC) in the urinary tract system depends on harboring virulence-associated genes encoded by mobile genetic materials such as the pathogenicity-associated Islands (PAIs) and bacterial plasmids (8). These virulence determinants play a critical role in UPEC pathogenicity by increasing their competitiveness and adaptability to the host urinary tract system (9). They enable the UPEC strains to colonize the urinary tract surfaces, invade the uroepithelial cells, evade the host defense mechanisms, and stimulate local inflammatory responses, causing various clinical manifestations (10).
FimH is an adhesin factor of type I fimbriae, expressed by over 90% of UPEC isolates (11). It provides an effective anchoring strategy, allowing the UPEC to adhere specifically to the surface of host cells, leading to colonization and invasion of bladder urothelium, causing lower UTIs (cystitis) (12). FimH represents the most abundant virulence gene among UPEC bacterial strains, revealing its crucial role in the pathogenesis of UPEC. For instance, the frequency of FimH was the highest compared to other virulence genes, with prevalence rates of 93%, 90%, 89%, 85%, and 68% in Romania, South India, Mongolia, Iran, and Tunisia, respectively (13-17).
Moreover, recent reports have shown a correlation between biofilm formation and the expression of the FimH virulence factor in UPEC strains (18). Antibiotic resistance among UPEC strains is a public health concern and is widely associated with therapeutic failure, contributing to a dramatic increase in mortality rates due to UTI-associated health complications (19). The resistance rate among uropathogens is significantly increasing, particularly in healthcare settings, due to the emergence of multidrug-resistant (MDR) organisms. These include carbapenem- resistant Enterobacteriaceae (CRE), extended-spectrum β-lactamase (ESBL)-producing organisms, and AmpC β-lactamase (20).
2. Objectives
In Lebanon, the epidemiological data that characterize the adhesion properties of UPEC clinical isolates is scarce. Thus, this study aimed to determine the prevalence of the UPEC adhesin gene (FimH), which plays a critical role in promoting the virulence properties of UPEC, and to characterize the resistance profile of UPEC by phenotypic screening of ESBL and MDR-producing UPEC isolates.
3. Methods
3.1. Study Design, Setting, and Population
A cross-sectional study was conducted on patients with UTIs in North Lebanon from July to September 2020. A total of 881 urine samples were collected from UTI patients admitted to different hospitals and private laboratories in Tripoli (Monla Hospital, Dar Al-Shifae Hospital, Middle East Medical Laboratory, and Al Mina Medical Lab), Koura (Al Koura Hospital), and Miniyeh-Dannnieh (El Kheir Hospital).
The processing of urine samples was conducted at different hospitals, and the pure cultures of UPEC isolates were collected and transferred under aseptic conditions to the Biomedical Laboratory of Beirut Arab University for further analysis. Patients with UTIs in the community or presenting as inpatients or outpatients in hospitals were included in our investigation.
3.2. Sample Collection and Processing
3.2.1. Urine Samples
Freshly voided midstream urine samples (10 - 20 mL) were collected into labeled, leak-proof, sterile containers at hospitals and private laboratories in North Lebanon. A standardized questionnaire was used to collect demographic characteristics and clinical information from each participant.
Initial urinalysis, including macroscopic and microscopic examinations and dipstick tests, was performed as a screening method to examine and evaluate the urinary tract disorders. The screening testing was followed by the standard urine culture technique to confirm the diagnosis of UTIs and identify the causative agents. Briefly, urine specimens of 10 μL were inoculated into MacConkey and chromogenic UTI agar plates using a sterile calibrated wire loop (0.001 mL). All plates were incubated at 37°C for 48 hours for visible growth and classified as significant, non-significant, or contaminated. At hospitals, bacterial colonies with significant growth (105 CFU/mL) on agar plates were identified using colony morphology, gram staining, and standard biochemical tests. The API 20E system was used for the confirmation of bacterial identity. After identification, the isolated UPEC strains were collected, labeled, and transported aseptically to the Biomedical Laboratory of Beirut Arab University for further analysis.
3.2.2. Uropathogenic <i>Escherichia coli</i> Isolates
The UPEC identification was further confirmed by VITEK® MS (BioMérieux, France), an automated microbial identification system, using matrix-assisted laser desorption deionization time of flight (MALDI-TOF) technology.
3.3. Antimicrobial Susceptibility Testing
The antimicrobial susceptibility testing of E. coli isolates was performed according to the Clinical and Laboratory Standards Institute (CLSI) guidelines using Kirby Bauer's disc diffusion method on Muller-Hilton agar.
A loopful (3 - 5 pure colonies) of freshly grown bacteria was suspended in 5 mL sterile saline solution and mixed gently to form a homogenous suspension. The turbidity of the bacterial suspension was adjusted to the density of a McFarland 0.5 standard. The obtained bacterial inoculums were streaked onto the surface of Mueller-Hinton agar plates using a sterile cotton swab.
Commercially available antibiotic disks of known concentrations were placed on the plates to evaluate the antimicrobial susceptibility profile of the selected isolates. They included amikacin (AN/ 30 μg), ampicillin (AMP/ 10 μg), augmentin (AMC/ 30 μg), aztreonam (ATM/ 30 μg), amoxicillin (AMX/ 20 μg), ceftriaxone (CRO/ 30 μg), ceftazidime (CAZ/ 30 μg), ciprofloxacin (CP/ 5 μg), cefepime (FEP/ 30 μg), cefixime (CFM/ 5 μg), cefotaxime (CTX/ 30 μg), cefoxitin (FOX/ 30 μg ), cefalotin (CEF/ 30 μg), cefuroxime (CXM/ 30 μg ), cefaclor (CEC/ 30), colistin (CST/ 50 μg), ertapenem (ETP/ 10 μg), fosfomycin (FOS/ 200 μg), gentamycin (GM/ 10 μg ), imipenem (IMP/ 10 μg), levofloxacin (LVX/ 5 μg), meropenem (MEM/ 10 μg), norfloxacin (NOR/ 10 μg), Nalidixic Acid (NA/ 30 μg), nitrofurantoin (F/ 300 μg), ofloxacin (OFX/ 5 μg), piperacillin (PRL/ 10 μg), piperacillin+ tazobactam (PPT 100/ 10 μg), ticarcillin (TIC/ 75 μg), ticarcillin/clavulanic acid (TCC) (75/ 10 μg), trimethoprim/sulfamethoxazole (SXT 1.25 μg/ 23.75 μg), tobramycin (TB/ 10 μg), and tetracycline (TET/ 30 μg).
After incubation at 37º C for 16 - 18 h, the diameter of the inhibition zone was measured, and the results were reported as sensitive (S), intermediate (I), or resistant (R). The isolates showing non-susceptibility to at least one antimicrobial agent in three or more antimicrobial categories were classified as MDR organisms. The UPEC isolates were further tested for ESBL by screening and confirmatory methods using the double-disk synergy test (DDST).
3.4. Quality Control
The sterility and performance of culture media were assessed before laboratory examinations, and standard reference strains of E. coli (ATCC 25922) were used to validate the culture and antimicrobial susceptibility testing results.
3.5. Phenotypic Detection of Extended-spectrum Beta-lactamase (ESBL)
3.5.1. Disc Susceptibility Test: Screening Assay
All UPEC isolates were screened for ESBL production using three cephalosporin indicators, including ceftazidime (30 μg), cefotaxime (30 μg), and cefpodoxime (30 μg). The UPEC isolates were regarded as resistant to these antimicrobial agents if the diameter of the inhibition zone for ceftazidime, cefotaxime, or cefpodoxime was ≤ 22 mm, ≤ 27 mm, or ≤ 17 mm, respectively. The UPEC strains, which were resistant to at least one of the three cephalosporins, underwent further phenotypic confirmation methods.
3.5.2. Double-disc Synergy Test (DDST): Confirmatory Method
To detect the ESBL-producing UPEC strains, antibiotic disks of ceftazidime (30 μg) and ceftriaxone (30 μg) were placed on Mueller-Hinton agar plates inoculated with the tested isolates at a distance of 20 mm from the combined disks of ceftazidime/clavulanic acid (30/10 μg) and ceftriaxone/clavulanic acid (30/10 μg). The plates were incubated overnight at 37°C. Any enhancement in the diameter of the inhibition zone (more than or equal to a 5 mm increase) for cephalosporins tested with clavulanic acid (CA) compared to its inhibition zone when tested alone was considered a positive ESBL result.
3.6. Genomic DNA Extraction
The UPEC genomic DNA was extracted using the NucleoSpin DNA extraction kit (Macherey-Nagel, Germany) according to the manufacturer's protocol.
3.7. Detection of the Uropathogenic <i>Escherichia coli</i> Isolates Adhesin Gene
The conventional polymerase chain reaction (PCR) assay was used to assess the prevalence of the UPEC adhesin gene (FimH) using specific primers (21).
3.8. Statistical Analysis
The statistical analysis was performed using IBM Statistical Packages for Social Sciences (IBM SPSS, version 22.00, IBM Corp., Armonk, N.Y., USA). Fisher's exact and t-tests were used to analyze the association between different variables. The P-value < 0.05 was considered statistically significant.
4. Results
4.1. Prevalence of UTIs and General Characteristics of Patients
Over a three-month period, 881 urine samples were collected from UTI-symptomatic patients admitted to different hospitals and private laboratories in Tripoli, Koura, and Miniyeh-Dannieh. Three hundred seventy urine specimens yielded significant bacterial growth (at least 105 CFU/mL), defining an infection rate of 42% (370/881). Among them, 19% (70/370) were identified as UPEC strains.
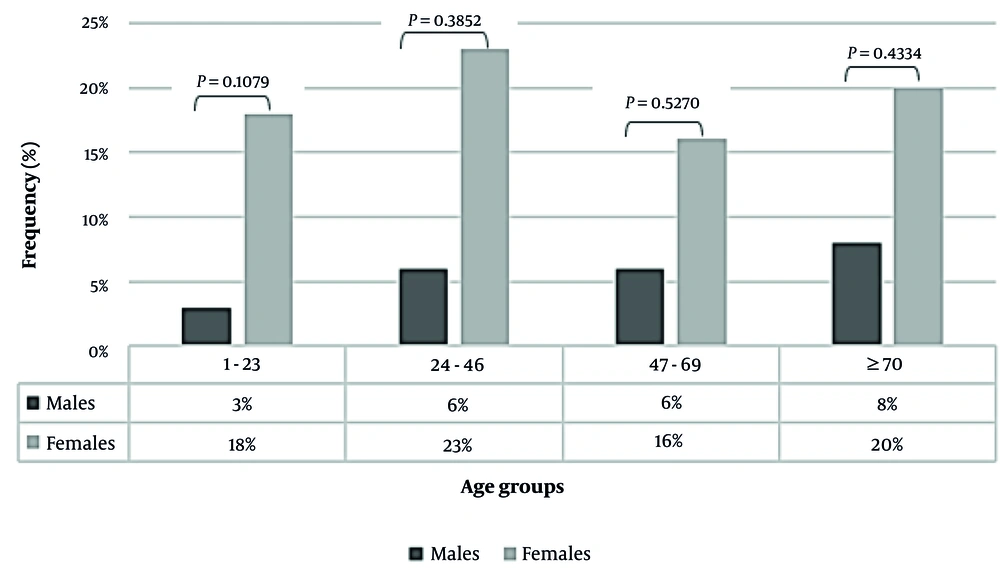
Of 70 UPEC-positive patients, females (77%; 54/70) had a higher UPEC infection rate than males (23%; 16/70). The age of these patients ranged between 1 and 96 years old, with a mean of 46 years old. Females belonging to age groups 24 - 46 (16/70; 23%) and above 70 years old (14/70; 20%) were most affected by UPEC strains compared to the men's group in all age categories (Figure 1).
4.2. Antibiogram Profile Among UPEC Isolates
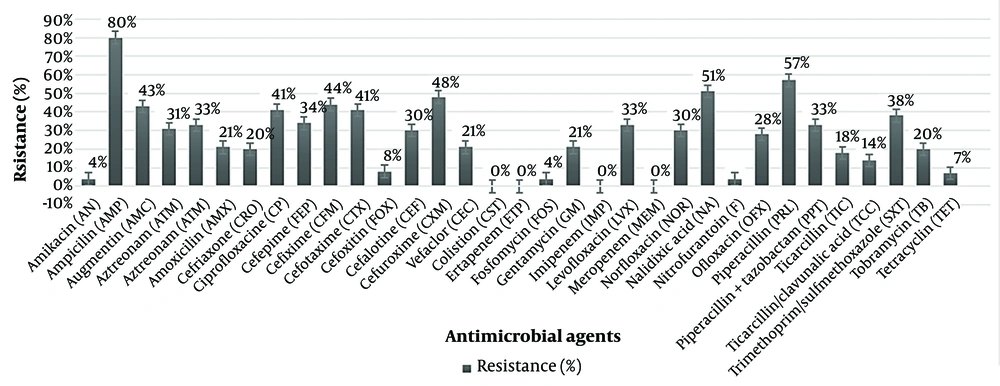
The antimicrobial susceptibility profile of all UPEC isolates was analyzed, and the resistance pattern is presented in Figure 2.
The highest resistance level was observed against ampicillin (80%; 56/70), followed by piperacillin (57%; 40/70), nalidixic acid (51%; 36/70), cefuroxime (48%; 34/70), cefixime (44%; 31/70), augmentin (43%; 30/70), cefotaxime (41%; 29/70), ciprofloxacin (41%; 29/70), trimethoprim/sulfamethoxazole (38%; 27/70), cefepime (34%; 24/70), piperacillin + tazobactam (33%; 23/70), amoxicillin (33%; 23/70), and levofloxacin (33%; 23/70). Maximum sensitivity among UPEC isolates was recorded for amikacin, colistin, ertapenem, imipenem, meropenem, nitrofurantoin, and fosfomycin.
4.3. Virulence Traits Among UPEC Isolates
The PCR assay revealed that 98.6% (69/70) of UPEC isolates expressed the FimH adhesin gene with a 419 bp amplicon size. The association between the expression of the FimH gene and the degree of resistance to antimicrobial agents among UPEC strains was also investigated (Table 1).
| Antibiotics Resistance (%) | FimH Positive (69/70), No. (%) | FimH Negative (1/70) (%) | P Value a |
|---|---|---|---|
| Amikacin (AN) | 2 (4) | 0 | 1 |
| Ampicillin (AMP) | 56 (80) | 0 | < 0.00001 b |
| Augmentin (AMC) | 29 (41) | 1 | < 0.00001 b |
| Aztreonam (ATM) | 21 (30) | 1 | < 0.00001 b |
| Amoxicillin (AMX) | 23 (33) | 0 | < 0.00001 b |
| Ceftriaxone (CRO) | 15, (21) | 0 | < 0.00001 b |
| Ceftazidime (CAZ) | 14 (20) | 0 | 0 b |
| Ciprofloxacin (CP) | 29 (41) | 0 | < 0.00001 b |
| Cefepime (FEP) | 24 (34) | 0 | < 0.00001 b |
| Cefixime (CFM) | 31 (44) | 0 | < 0.00001 b |
| Cefotaxime (CTX) | 29 (41) | 0 | < 0.00001 b |
| Cefoxitin (FOX) | 6 (8) | 0 | 0.0099 b |
| Cefazolin (CEF) | 21 (30) | 0 | < 0.00001 b |
| Cefuroxime (CXM) | 34 (48) | 0 | < 0.00001 b |
| Cefaclor (CEC) | 14 (20) | 1 | < 0.00001 b |
| Colistin (CST) | 0 (0) | 0 | 1 |
| Ertapenem (ETP) | 0 (0) | 0 | 1 |
| Fosfomycin (FOS) | 3 (4) | 0 | 1 |
| Gentamycin (GM) | 15 (21) | 0 | 0.0001 b |
| Imipenem (IMP) | 0 (0) | 0 | 1 |
| Levofloxacin (LVX) | 23 (33) | 0 | < 0.00001 b |
| Meropenem (MEM) | 0 (0) | 0 | 1 |
| Norfloxacin (NOR) | 21 (30) | 0 | < 0.00001 b |
| Nalidixic acid (NA) | 35 (50) | 1 | < 0.00001 b |
| Nitrofurantoin (F) | 3 (4) | 0 | 1 |
| Ofloxacin (OFX) | 20 (28) | 0 | < 0.00001 b |
| Piperacillin (PRL) | 40 (57) | 0 | < 0.00001 b |
| Piperacillin+ tazobactam (PPT) | 23 (33) | 0 | < 0.00001 b |
| Ticarcillin (TIC) | 13 (18) | 0 | < 0.00001 b |
| Ticarcillin/clavulanic acid (TCC) | 10 (14) | 0 | < 0.00001 b |
| Trimethoprim/sulfamethoxazole (SXT) | 26 (37) | 1 | < 0.00001 b |
| Tobramycin (TB) | 14 (20) | 0 | 0.0001 b |
| Tetracycline (TET) | 5 (7) | 0 | 0.0002 b |
Distribution of Antibiotic Resistance Among Uropathogenic Escherichia coli Strains and its Association with FimH
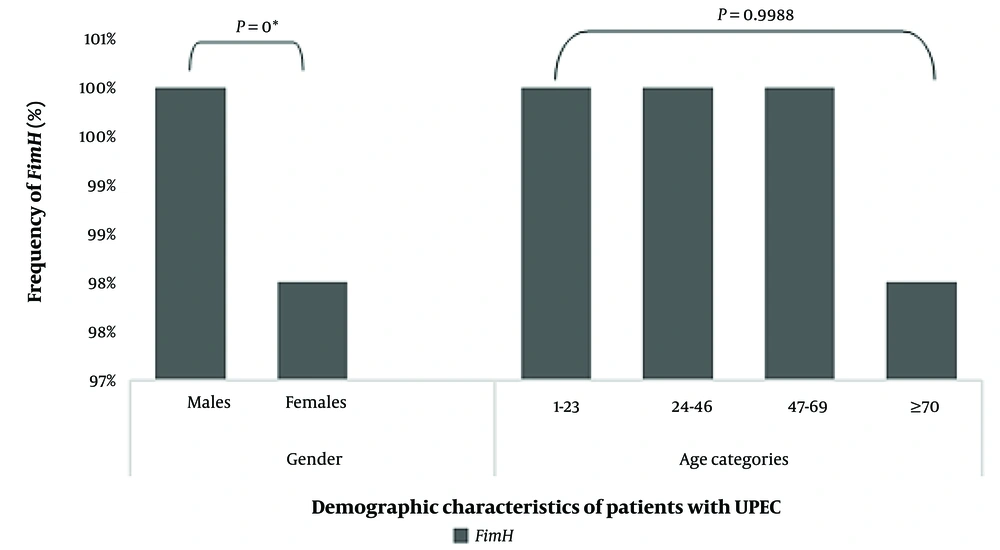
A significant association was found among UPEC isolates between FimH production and resistance to all tested antimicrobial agents (P < 0.05), except for amikacin (AN), ceftazidime (CAZ), colistin (CST), ertapenem (ETP), fosfomycin (FOS), imipenem (IMP), meropenem (MEM), nitrofurantoin (F), and tazobactam (TZP). The prevalence of the FimH adhesin gene among UPEC isolates was also assessed concerning the gender and age groups of UTI patients (Figure 3).
Demographic characteristics of patients with uropathogenic Escherichia coli (UPEC). * Statistically significant P values are calculated by Fisher's exact test (association between FimH expression among UPEC isolates and gender) and t-test (association between FimH production and different age groups of UTI patients).
Also, FimH expression was higher in males than in females (P = 0). However, no significant differences were reported among different age groups (P = 0.9988).
4.4. Distribution of ESBL and MDR Among UPEC
According to Magiorakos' criteria, which define MDR as acquired resistance to at least one antibiotic agent in three or more antimicrobial classes (22), approximately 68% (48/70) of UPEC isolates were MDR, of which 98% (n = 47/48) expressed the FimH adhesin gene.
The laboratory confirmatory test revealed that 64% (45/70) of the tested UPEC strains were phenotypically identified as extended-spectrum beta-lactamase-producing organisms, of which 100% (n = 45) expressed the FimH adhesin gene.
5. Discussion
The pathogenicity of UPEC in UTIs depends on its virulence properties, which affect the degree of colonization, invasion, and proliferation within the umbrella cell of the urothelium (10). Adhesion, the initial stage of the UPEC infection cycle, is considered the most critical step, which determines the ability of UPEC strains to cross the uroepithelial cells and invade deeper tissues, initiating the proliferation and formation of an intracellular bacterial community (IBC) (12).
As known, UPEC attachment to uroepithelial cells is mediated by different adhesin factors, including FimH, which is located at the tip of type 1 fimbriae and plays a critical role in protecting the pathogens from the clearance effects of urine flow (23). The characterization of UPEC virulence traits and their correlation with the prevalence of antibiotic resistance is not well studied in Lebanon. Thus, getting insights into UPEC virulence factors will help develop novel alternative therapies for UTIs by designing selective FimH antagonist drugs that can abolish the progression of UTIs without resorting to long-term antimicrobial therapy, which has detrimental effects on the normal configuration of the gut microbiota (24). The current investigation reported a prevalence of 42% (370/881) of UTIs in patients with urinary tract discomfort residing in North Lebanon, of which 19% (70/370) were caused by UPEC organisms.
A study conducted in South Lebanon on a total of 551 UTI patients of all age groups attending medical laboratories or urology departments showed a lower prevalence of UTIs (22.1%; 682/3082) but a higher rate of UPEC isolates (67.1%; 449/682) compared to our findings (21). In contrast, a study conducted in Egypt reported a higher prevalence of UTIs, accounting for 68.6% (400/583), and a greater UPEC isolation rate of 33.5% (134/400) (25).
Our study showed that females are at higher risk of developing UTIs than males, with a prevalence rate of 77% (54/70). The high frequency of UTIs among women is mainly attributable to the anatomical structure of the genitourinary tract, which favors bacterial colonization and invasion (26). In addition, our study revealed that females aged 24 - 46 years and those older than 70 years were predominantly affected by UPEC organisms, with prevalence rates of 23% (16/70) and 20% (14/70), respectively. These findings are supported by a study conducted in Uganda (27).
Older women are more susceptible to UTIs due to age-associated factors, reduced immune system activity, high exposure to nosocomial uropathogens, and rising health comorbidities (28). The antimicrobial susceptibility testing has revealed that UPEC strains showed a high degree of resistance to the beta-lactam class of antibiotics, including piperacillin and piperacillin-tazobactam. This was in line with a study conducted in Iran, where a similar resistance level (55.8%) against piperacillin was reported (7). Conversely, a minimal resistance pattern against piperacillin-tazobactam was recorded in Romania and Africa, with resistance levels of 5.97% and 0%, respectively (14, 29).
In addition, our results reported the highest resistance degree among UPEC strains against the second-generation cephalosporin (Cefuroxime; 48%; 34/70), followed by the third and fourth-generation cephalosporins (cefixime, cefotaxime, and cefepime) with resistance levels of 44% (31/70), 41% (29/70), and 34% (24/70), respectively. A study conducted in Columbia reported higher resistance levels to cefepime (54.2%) and cefotaxime (44.2%), whereas the resistance degree to ceftazidime and ceftriaxone was lower compared to our findings with reported levels of 11.1% and 5.3%, respectively (30).
In our study, the highest resistance level among UPEC was observed against the therapeutic agents that are commonly used as first-line empirical treatments for UTIs, including ampicillin, nalidixic acid, augmentin, ciprofloxacin, and sulfamethoxazole-trimethoprim. These rates were remarkably lower than the resistance levels reported in Iran against ampicillin (88.9%), nalidixic acid (51%), and ciprofloxacin (56.6%) (31). The growing resistance pattern against these antimicrobial agents may be due to the misuse of antibiotics, the long course of antibiotherapy, and the horizontal transfer mechanisms of resistant genes among UPEC organisms (7).
In this study, the tested UPEC isolates showed resistance to ciprofloxacin and levofloxacin, with reported levels of 41% and 33%, respectively. These results are consistent with those reported in Columbia (30) but higher than the prevalence rates observed in Romania (14). The majority of UPEC isolates in this study were susceptible to the clearance effects of amikacin and the carbapenem class of antibiotics (imipenem, ertapenem, and meropenem), as well as nitrofurantoin, colistin, and fosfomycin. This concorded with several studies (7, 14, 31).
The emergence of antibiotic resistance among UPEC organisms has restricted the incorporation of imipenem in the first-line therapy of UTIs, and its use has been limited to the treatment of ESBL-producing organisms (32).
Despite the effectiveness of nitrofurantoin to combat the UPEC strains, the clinical applications are limited due to various side effects. Nevertheless, the increasing resistance rates among UPEC pathogens against the first-line antimicrobial agents would suggest the rational use of nitrofurantoin for recurrent, uncomplicated UTIs (32).
In this study, the frequency of ESBL-producing UPEC isolates was 64% (45/70). This was in line with another study conducted in Egypt showing a similar prevalence of ESBL among the examined UPEC organisms (25). In our study, most UPEC isolates, 68% (48/70) showed considerable resistance to almost all tested antimicrobial agents and were classified as MDR. This was in line with other studies conducted in Nepal and Iran, where the prevalence rates of MDR ranged between 51% and 79% (7, 31). Molecular analyses showed that almost all the tested UPEC isolates (98.6%; n = 69) harbored the FimH adhesin gene. Similar prevalence rates were reported in other studies in India, Romania, and Pakistan (13, 14, 33). However, lower incidence rates of FimH were reported in Iran (34.1%) (34).
Our results emulated a significant correlation (P < 0.05) between FimH and the emergence of resistance against the majority of the tested antimicrobial agents (Table 1). Similar findings were reported, showing a significant association (P < 0.05) between MSHA (type 1 fimbriae) and resistance to antimicrobial agents among UPEC strains (26). However, a study conducted in Iran has negated this association (5).
Our study found a significant association between the UPEC virulence gene (FimH) and the gender of UTI patients (Figure 3). This was in contrast to a study conducted in Iran, where no significant difference was found in the prevalence of FimH concerning the participants' genders (14). However, this report confirmed our finding regarding the absence of a relationship between FimH-producing UPEC and different age categories of UTI patients.
An association between FimH expression among UPEC strains and the emergence of MDR organisms was reported in our study (P < 0.05), which was in line with an Iranian report (35).
All the ESBL-producing organisms in this study harbored the FimH adhesin gene, revealing a statistically significant association (P < 0.05) between the ESBL phenotype and FimH among UPEC isolates. However, a study conducted in Iran did not support such an association (35).
FimH is one of the most important adhesin factors encoded by UPEC pathogens. It plays a critical role in mediating the UPEC-target cells' stereochemical interactions to enhance the adhesion and colonization of bacteria on the urothelium. Furthermore, FimH mediates the mechanism of biofilm formation and the quorum sensing strategy between invading bacterial cells, resulting in an increase in virulence in UPEC organisms. The virulence-enhancing properties of FimH could explain our main findings, which suggest a possible association between the increased antimicrobial resistance and the presence of the adhesin gene, as well as between FimH expression and the incidence of MDR and ESBL among UPEC strains.
5.1. Conclusions
The prevalence of UTIs in North Lebanon was determined in this study, and it reached 42%, with UPEC representing 19% of the detected uropathogens. The antimicrobial susceptibility testing showed that about 68% (n = 48) of the tested UPEC were multidrug-resistant (MDR), and 98% (n = 47) of the studied isolates were phenotypically confirmed as ESBL-positive strains. The molecular analysis revealed that most UPEC strains (98.6%; n = 69) harbored the FimH adhesin gene. In addition, a significant association was found between the FimH adhesin factor and the emergence of resistance among UPEC strains (P < 0.05).
Overall, the present study provides relevant epidemiological data characterizing the UPEC prevalence and virulence properties in Lebanon. Moreover, these findings enhance our understanding of the role of FimH in the pathogenesis of UPEC, which in turn could help develop novel alternative therapies for managing UTIs and thus decrease the inadequate administration of antimicrobial agents.