1. Background
UTIs are commonly caused by Gram-negative bacteria. E. coli is responsible for more than 80% of the UTIs (1). Broad-spectrum cephalosporins such as cefotaxime, ceftazidime and cefepime are among the most important antibiotics to treat infections caused by Gram-negative bacilli (2). Gram-negative bacilli such as E. coli become resistant to antibiotics through several mechanisms, including production of β-lactamase enzymes, changes in penicillin-binding proteins (PBPs), drug permeability reduction through mutations in porins and increase of the activity of efflux pumps (2, 3). Production of β-lactamase enzymes are among the most common mechanisms of resistance to β-lactam antibiotics in E. coli (2, 3). Several phenotypes of β-lactamase enzymes including extended spectrum β-lactamases (ESBLs), metallo-β-lactamases (MBLs) and Amp-C β-lactamases have reported in Gram-negative bacilli such as Enterobacteriaceae family, Pseudomonas aeruginosa (P. aeruginosa) and Acinetobacter spp. (3). There are several methods to identify ESBLs (3, 4). Combination double disk synergy test (CDDST) is a confirmatory method that can approve the detection of ESBLs in Gram-negative bacilli such as Enterobacteriaceae (4). Nowadays, due to outbreaks of isolates with multiple mechanisms of resistance to β-lactam antibiotics, sensitivity of CDDST method to detect ESBLs producing Gram-negative bacilli has decreased because of overlapping between the resistance mechanisms (5, 6). The simultaneous presence of multiple resistance mechanisms such as producing of ESBLs, AmpC, mutations in porins, lack of drug penetration into the bacterial cell and efflux pumps activity increase, may overlap with each other (7-9). Therefore, it is essential to introduce a method capable to detect ESBLs in Gram-negative bacilli with resistance to β-lactam antibiotics through multiple resistance mechanisms simultaneously. β-lactamase enzymes are secreted to periplasmic space of Gram-negative bacilli after synthesis (7-9). β-lactamase disk test is a modified method to detect ESBLs in Gram-negative bacilli. In this method, after the destruction of the outer membrane by Tris-EDTA (TE) buffer, ESBLs are released into the medium culture and cause the hydrolysis of β-lactam antibiotics (9, 10). Outer membrane destruction leads to omission of other β-lactam resistance mechanisms such as activity of efflux pumps and lack of drug penetration (6).
2. Objectives
The current study aimed to evaluate the ability of the CDDST and β-lactamase disk test to detect ESBLs in clinical isolates of E. coli resistant to broad-spectrum cephalosporins and introduce a new modified method to detect ESBLs.
3. Methods
3.1. Bacterial Isolates
The current experimental study collected 86 MDR E. coli isolates from June 2014 to March 2015, from the urine sample of different patients demonstrating symptoms of UTIs admitted to hospitals of Kerman University of Medical Sciences. All isolates were considered as E. coli by standard biochemical tests including: oxidase test, lactose fermentation, growth on triple sugar iron medium, Simmons’ citrate agar and MRVP (methyl red-Voges Proskauer) broth.
3.2. Susceptibility Testing
Standard disk diffusion method on Mueller Hinton agar (CONDA Co; Spain) was used to determine antibiotic resistance profile according to Clinical and Laboratory Standards Institute (CLSI) recommendations (10). Antibiotic disks (HiMedia Co; India) included included: amoxicillin/clavulanic acid (10 µg/20 µg), cefotaxime (30 μg), ceftazidime (30 μg), cefepime (30 μg), imipenem (10 μg), gentamicin (10 μg), ciprofloxacin (5 μg) and trimethoprim/sulfamethoxazole (1.25 µg/23.75 µg). E. coli strain ATCC 25922 was used as standard strain in disk diffusion method (11). The isolates resistant to at least three antibiotics from different classes were considered as MDR phenotypes according to the recommendations by Magiorakos et al. (12).
3.3. Identification of ESBLs Producing Isolates by CDDST
To perform this procedure, 0.5 McFarland standard was prepared from the clinical isolates and then cultivated on Mueller-Hinton agar (CONDA Co; Spain). On Mueller-Hinton agar the disks were placed as follows: ceftazidime (30 μg), cefotaxime (30 μg), cefpodoxime disks alone, and plus clavulanic acid (10 μg); they were placed at a distance of 20 mm from each other and then incubated overnight at 37°C. Isolates that their growth inhibition zone diameter around the clavulanic acid containing disks, were ≥ 5 mm of the non-clavulanic acid containing disks, considered as ESBLs producing isolates. In this method, Klebsiella pneumoniae (K. pneumoniae) ATCC700603 was used as positive control and E. coli ATCC 25922 as a negative control (4).
3.4. Identification of ESBLs-Producing Isolates by β -lactamase Disk Test
The design of this test was based on the presence of ESBLs in periplasmic space of Gram-negative bacilli (7-9). In this method, briefly, 100X TE buffer was diluted with normal saline in equal proportions (1/1). Then 20 mL of this solution was added to the blank disks (no antibiotic) and used after drying at room temperature. Then the cefotaxime sensitive P. aeruginosa ATCC 27853 (tested by 30 µg disk) was cultured on Mueller-Hinton agar. A cefotaxime disk (30 µg) was placed on the plate and two TE buffer-containing blank disks were placed on the top and bottom of it. Then colonies of the clinical isolates were added to each of the TE buffer containing disks. The TE buffer on the disk causes destruction in the outer membrane and leads to release β-lactamase enzymes into the culture medium. Flattening or indentation of the growth inhibition zone of the CTX disk at the side of blank disks containing the test isolates indicated that the release of ESBLs leads to cefotaxime sensitive P. aeroginosa growth toward cefotaxime disk in the plate (9, 10, 13). P. aeruginosa ATCC 27853 and E. coli ATCC 25922 were used as negative controls in this method (4).
3.5. Detection and Sequencing of β-lactamase Genes by PCR
To identify β-lactamase genes, bacterial DNA was extracted by boiling method. The genes blaTEM, blaSHV, blaOXA, blaPER-1 and blaCTX-M ESBL were identified, using specific primers listed in Table 1. The PCR was carried out in FlexCycler PCR Thermal Cycler (Analytik Jena, Germany) by Red PCR Master mix (Ampliqon Co, Denmark) according to the manufacture’s instruction, under the following conditions: initial denaturation in 95°C for five minutes followed by 30 cycles including: denaturation in 95°C for one minute, annealing for one minute (temperatures are listed in Table 1), extension in 72°C for one minute and the final extension in 72°C for five minutes. PCR reactions were performed in total volume of 25 µl. The master mix contained 12.5 µL of reaction mixture containing Taq DNA Polymerase, 0.5 µl of each forward and reverse primers (10 pM), 2 µl of target DNA (20 ng) and 9.5 µl of distilled water. The PCR products were electrophoresed on 1.2% agarose gel (Sigma, USA) containing Green viewer dye (Pars Tous, Iran), and then the gel was visualized by a Gel Doc TM XR image analysis station (Bio-Rad, Hercules, USA). PCR products were purified for sequencing after separation on an agarose gel using the GF-1 PCR Clean up Kit (VIVANTIS Inc., Malaysia) following the manufacturer’s instructions. Sequences were determined, assembled and compared with the sequences in the GenBank. K. pneumoniae ATCC700603 (positive for blaTEM and blaSHV) and P. aeruginosa strain KOAS (positive for blaPER) were used as control positive in PCR reaction.
| Gene Target | Primer Sequences (5' - 3') | Annealing Temperature (°C) | Product Size (bp) | Reference |
|---|---|---|---|---|
| blaSHV | F-TCAGCGAAAAACACCTTG | 56 | 472 | (4) |
| R-TCCCGCAGATAAATCACC | ||||
| blaTEM | F-CTTCCTGTTTTTGCTCAC C | 56 | 636 | (4) |
| R-AGCAATAAACCAGCCAGC | ||||
| blaPER | F-ATGAATGTCATTATAAAAGC | 49 | 925 | (13) |
| R-AATTTGGGCTTAGGGCAGAA | ||||
| blaOXA | F-TCAACTTTCAAGATCGCA | 49 | 601 | (14) |
| R-GTGTGTTTAGAATGGTGA | ||||
| blaCTX-M | F-CGCTTTGCGATGTGCAG | 49 | 550 | (15) |
| R- ACCGCGATATCGTTGGT |
3.6. Statistics Analysis
The sensitivity and specificity of phenotypic methods compared with those of PCR were analyzed by medCalc 81, available online (https://www.medcalc.org/calc/diagnostic_test.php).
4. Results
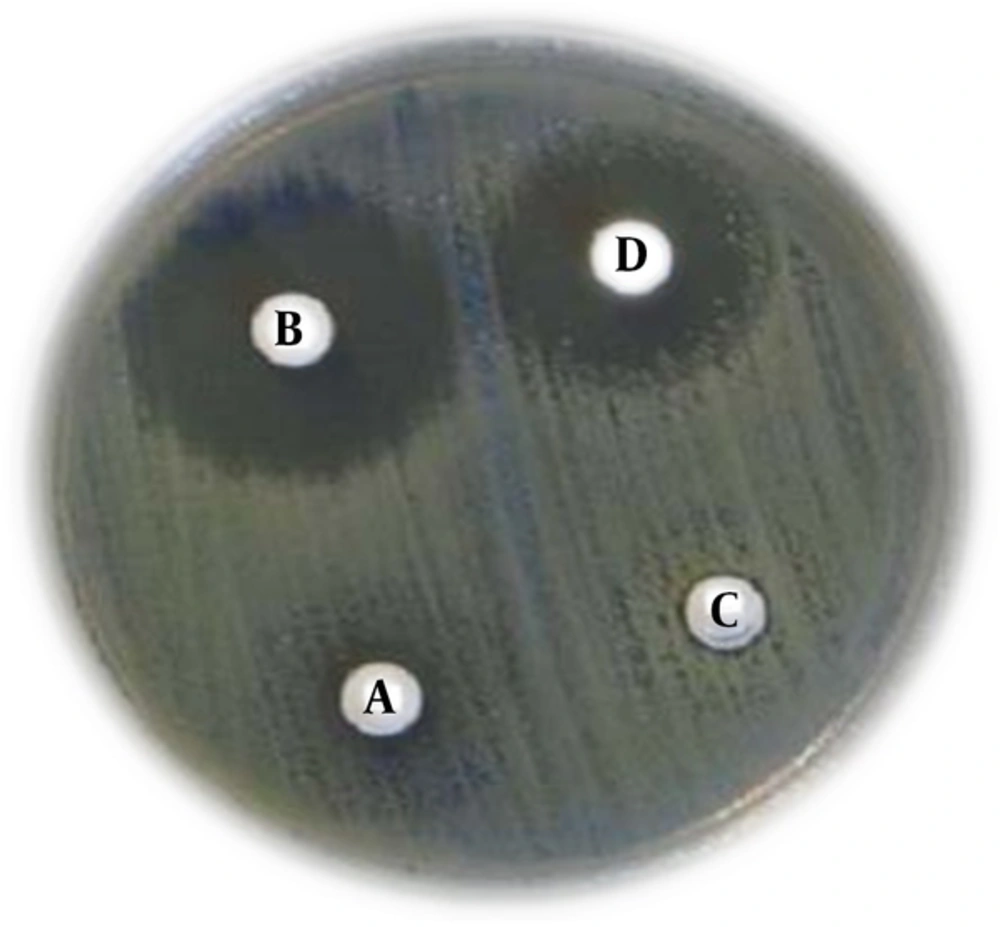
Among the 86 isolates, antibiotics resistance profile was as follows: cefotaxime (96.5%), ceftazidime (91.8%), cefepime (88.3%), amoxicillin/clavulanic acid (81.3%), ciprofloxacin (84.8%), trimethoprim/sulfamethoxazole (87.2%), gentamicin (32.5%) and nalidixic acid (93%). All isolates were sensitive to imipenem. All isolates were resistant to more than three antibiotics of different classes; therefore, they all were considered - as MDR phenotype. 88% and 97.6% of the isolates were considered as ESBLs producing by CDDST and β-lactamase disk tests, respectively (Figures 1 and 2). In CDDST, eight isolates were detected with no growth inhibition zones around cefotaxime, ceftazidime and cefpodoxime disks alone, and plus clavulanic acid and according to CLSI guidelines for CDDST were considered as negative for ESBLs production, however these eight isolates were positive for ESBLs genes (Table 2). All these eight isolates hydrolyzed cefotaxime in β-lactamase disk test and were considered ESBLs producing. Fifty-three (61.6%) isolates were positive for blaTEM, 64 (74.4%) for blaCTX-M, 12 (14%) for blaOXA-1 and 2 (2.3%) for blaSHV genes. All isolates were negative for blaPER gene. Also 92% of the isolates (79 isolates) had at least one of the ESBLs genes including blaTEM, blaCTX-M, blaOXA-1 and blaSHV. Co-existence rate of the ESBLs genes was as follows: 6 (7%) for blaOXA-1, blaCTX-M and blaTEM, 1 (1.1%) for blaSHV, blaCTX-M and blaTEM, 36 (41.8%) for blaTEM and blaCTX-M, 3 (3.5%) for blaTEM and blaOXA-1, 1 (1.1%) for blaSHV and blaCTX-M and 2 (2.3%) for blaOXA-1 and blaCTX-M. PCR products were sequenced and then deposited into the GenBank under the following accession numbers: KU059762 (blaSHV), KU059763 (blaTEM), KX168501 (blaCTX-M) and KU059764 (blaOXA). Sensitivity, specificity, positive predictive value and negative predictive value of CDDST and β-lactamas disk test are shown in the Table 3.
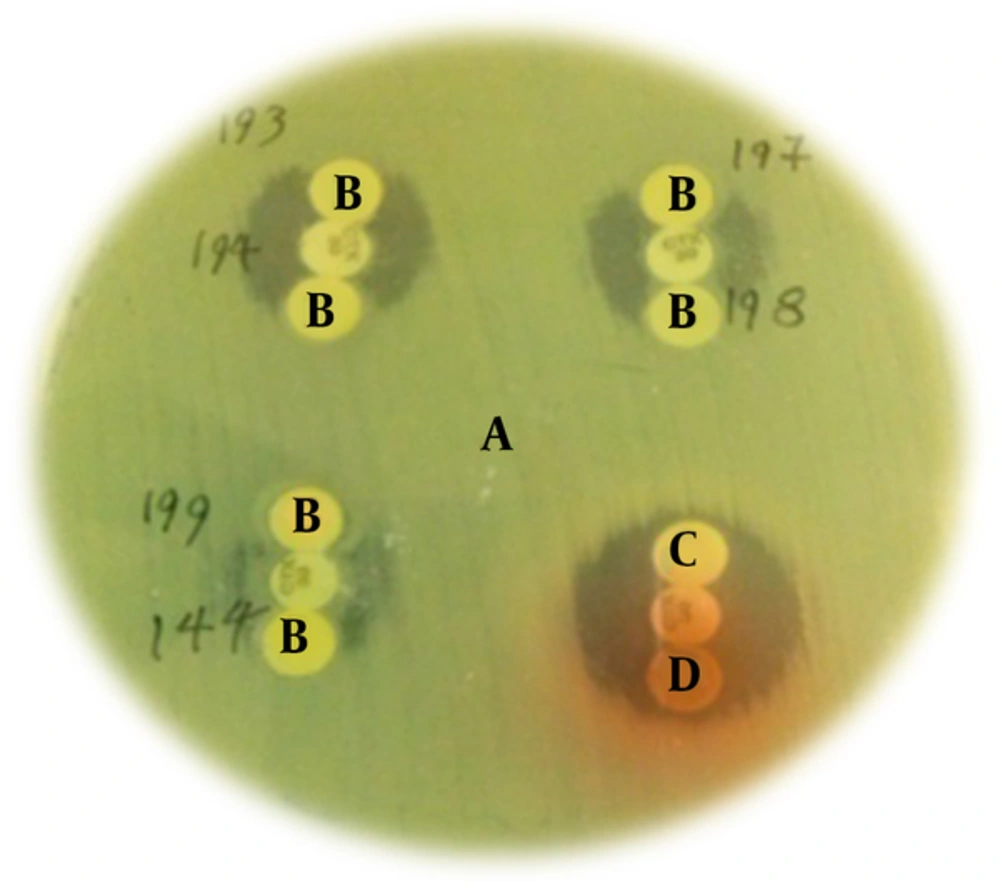
β-lactamase disk test; A: (Lawn culture) P. aeroginosa ATCC 27853, B: (TE buffer containing blank disk, dipped with clinical isolates and cefotaxime disk in the middle) ESBLs producing clinical isolates, C & D: negative control; E.coli ATCC 25922 and P. aeroginosa ATCC 27853. ESBLs production by clinical isolates causes changes in the growth inhibition zone form (clover leaf figure) around the cefotaxime disk.
| Number of Isolates | CDDST | β-Lactamase Disk Test | blaTEM | blaTEM + blaCTX-M | blaTEM + blaOXA | blaTEM + blaOXA + blaCTX-M |
|---|---|---|---|---|---|---|
| 2 | - | 2 | 2 | - | - | - |
| 1 | - | 1 | - | - | 1 | - |
| 2 | - | 2 | - | - | - | 2 |
| 3 | - | 3 | - | 3 | - | - |
Abbreviation: CDDST, combination double disk synergy test.
aTotal = 8.
| Phenotypic Tests | Sensitivity (%) | Specificity (%) | Positive Predictive Value | Negative Predictive Value |
|---|---|---|---|---|
| CDDST | 90.59% | 100.00% | 100.00% | 89.74 % |
| β-lactamase disk test | 96.34% | 100.00 % | 100.00% | 95.89 |
Abbreviation: CDDST, combination double disk synergy test.
5. Discussion
According to the definitions (12), all isolates examined in the current study were MDR, which indicated the association between ESBLs production and resistance to other antibiotics such as fluoroquinolones, aminoglycosides and trimethoprim/sulfamethoxazole (2, 3).
In the current study, 88% and 97.6% of the isolates were identified as ESBLs producing by the CDDST and β-lactamase disk test, respectively. According to the PCR results, 92% of isolates had at least one of the ESBLs genes such as blaTEM, blaCTX-M, blaOXA-1 and blaSHV; these findings were close to β-lactamase disk test results. In this study and by the CDDST, eight of the isolates had no growth inhibition zone around cefotaxime, ceftazidime and cefpodoxime disk alone, and plus clavulanic acid and according to CLSI guidelines, due to the lack of inhibition zones around the disks containing clavulanic acid, were considered negative for ESBLs production (11). However all these eight isolates were considered ESBLs producing when β-lactamase disk test was used, due to hydrolysis of cefotaxime. This result may indicate the presence of multiple mechanisms of resistance to broad-spectrum cephalosporins in such isolates and when these mechanisms overlap, it leads to the reduction of CDDST sensitivity to identify the ESBLs producing isolates (3, 5). Several studies reported that CDDST was not suitable to detect ESBLs producing isolates. In previous studies in 2010 and 2012 in Kerman, Iran, E. coli and K. pneumoniae isolates with no growth inhibition zones around the cefotaxime, ceftazidime and cefpodoxime disks alone, and plus clavulanic acid , were reported (16-19). Also in other reports in 2014 and 2015 in Tehran, conducted on 100 carbapenem-resistant P. aeruginosa isolates that were positive for blaTEM, blaOXA and blaSHV, only three isolates were considered as ESBLs producing by CDDST method and 97 isolates exhibited no growth inhibition zones around the cefotaxime, ceftazidime and cefpodoxime disks, alone or in combination with clavulanic acid (5, 13). Also, Garrec et al., reported that in AmpC co-producers isolates CDDST method were not suitable to detect ESBLs (sensitivity = 46% - 85%) (20-22). These results, similar to those of the current study, indicated that the presence of multiple mechanisms could decrease the sensitivity of ESBLs detection by CDDST method (20-22). The results of the current study and other studies indicated the emergence of Gram-negative bacilli strains that simultaneously had several mechanisms of resistance to β-lactam antibiotics; leading to the reduction of CDDST sensitivity to identify ESBLs producing Gram-negative bacilli (5, 9, 13).
In the current study, eight isolates positive for β-lactamase genes were negative for ESBLs production when tested by the CDDST, but they were considered as ESBLs producing by β-lactamase disk test. Since in the β-lactamase disk test, the outer membrane is disrupted, positive results in the test could indicate the role of outer membrane associated drug resistance mechanisms such as active drug efflux pumps and the lack of penetration due to mutations in porins (3, 7). In summary, it was demonstrated that β-lactamase disk test had high sensitivity (sensitivity = 96.34%) to detect ESBLs in Gram-negative bacilli, particularly in isolates with several mechanisms of resistance to β-lactam antibiotics. Finally, since the β-lactamase disk test is a modified method of ESBLs detection therefore more studies are needed to confirm its ability to detect ESBLs among Gram-negative bacilli.
5.1.Conclusion
The β-lactamase disk test could be a particularly appropriate method to detect ESBLs producing isolates with resistance to β-lactam antibiotics through several mechanisms.