1. Background
World Health Organization (WHO) outlined multi-drug resistant Gram-negative bacteria (MDR-GNB) among the priority list of pathogens that pose the greatest threat to human health. Infection with these pathogens occurs in children with underlying diseases, leading to death as a common consequence of long-term hospitalization (1, 2). The rate of bloodstream infections by Gram-negative bacteria (GNB) has been increasing among vulnerable children in recent years, and there is a possibility of repeated bacteremia in immunodeficient patients (3). The GNB has a major responsibility for increasing bacteremia among children (4). Among GNB, extended-spectrum β-lactamase producing Enterobacteriaceae (ESBL-E), carbapenem-resistant Enterobacteriaceae (CRE), fluoroquinolone-resistant GNB, and Pseudomonas aeruginosa are the main therapeutic challenges for patients with bacteremia (5).
Several risk factors, including age, underlying diseases, chemotherapy regimens, previous history of hospitalization, length of hospital stays, and administration of broad-spectrum antibiotics, can cause bloodstream infection (BSI) (6). COVID-19 is one of the newly emerged pathogens for children and adults, caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Although several studies have been done on COVID-19 in children, its co-occurrence with bacteremia and its impact on the etiology of BSI, mainly through GNB, is not well-known in children. Although the secondary bacterial infection rate has not been high, broad-spectrum antibiotics are used as prophylaxis treatment for most patients with COVID-19, leading to the appearance of new MDR strains with new resistant mechanisms in both hospitals and communities (7, 8). Bacterial and viral superinfections have been introduced as a critical point since previous epidemics and pandemic outbreaks of viral respiratory infections, particularly in the influenza A virus subtype (H1N1) in 2009, with 30% of confection report in critically ill patients (9, 10). The recorded mortality of viral and bacterial superinfections was high in the last influenza pandemic, and GNB was the most frequent strain isolated from the patients (11). Gram-negative bacteria were the most prevalent pathogens in most patients who were admitted to the hospital. They were transferred to the intensive care unit (ICU) after a COVID-19 confirmation test that could lead to hospital-acquired pneumonia (HAP) or ICU-HAP (12). There is a gap in knowledge about specific data on antimicrobial resistance patterns in GNB isolated from patients with viral and bacterial co-infections (13).
2. Objectives
Due to the lack of data about concomitant infections with SARS-CoV-2, this study aimed to analyze the co-occurrence of COVID-19 and bacteremia in children. We also investigated the possible correlation between these two diseases, types of GNB-causing bacteremia, and their antimicrobial resistance phenotypes. This kind of study can be useful for a better understanding of concomitant infections in children.
3. Methods
3.1. Patients and Samples
Blood samples of hospitalized patients in a teaching hospital in Iran, with a clinical demonstration of BSI, were collected by Becton Dickinson Diagnostic Instrument Systems, Sparks, Md (BACTEC) bottles for 2 years from 2020 to 2022. The patients were admitted to different wards of the hospital, including oncology, bone marrow transplantation (BMT), surgery, neurology, rheumatology, pediatric intensive unit (PICU), infectious disease, emergency, and gastrointestinal patients up to 17 years old. All of the patients were tested for COVID-19 in the case of susceptive clinical demonstration, such as fever and cough, by the hospital laboratory. This study was approved by the Ethics Committee of Shahid Beheshti University of Medical Sciences (IR.SBMU.MSP.REC.1398.1041). Demographic information of the patients was collected via hospital documents, a questionnaire, which was signed by patients’ parents, and an informed consent form.
3.2. Anti-microbial Susceptibility Testing and Multi-drug Resistant Gram-negative Bacteria Characterization
The inoculated pediatric BACTEC bottles were followed up in the device for seven days until they reported a positive alarm. Moreover, 33 mL of blood samples were centrifuged for ten minutes at 4,000 rpm; then the supernatant was removed, and 25 µL of the sedimentation was cultured in conventional and differential culture media of blood agar and MacConkey agar at aerobic conditions in 37°C incubators. The growth of the colonies was investigated after 24 hours and screened for GNB. Fermentation of sugars, reduction of indole, motility, production of acetyl methyl carbinol using Voges Proskauer, use of citrate as sole carbon source, decarboxylation of amino acids, and urease activity were done as biochemical tests. The susceptibility and resistance of the strains were recorded for Enterobacteriaceae family members, Acinetobacter, and Stenotrophomonas spp. against gentamycin (10 µg), tobramycin (10 µg), amikacin (30 µg), ciprofloxacin (5 µg), levofloxacin (5 µg), piperacillin (30 µg), ceftazidime (30 µg), piperacillin-tazobactam (110 µg), cefepime (30 µg), aztreonam (30 µg), imipenem (10 µg), meropenem (10 µg), trimethoprim-sulfamethoxazole (2.5 µg), cefotaxime (30 µg), ampicillin (10 µg), ampicillin-sulbactam (20 µg), doxycycline (30 µg), doripenem (10 µg), tetracycline (30 µg), chloramphenicol (30 µg), cefuroxime (30 µg), and cephalexin (30 µg). All the following antibiotics were also tested for Pseudomonas spp. except trimethoprim-sulfamethoxazole (2.5 µg), cefotaxime (30 µg), ampicillin (10 µg), ampicillin-sulbactam (20 µg), doxycycline (30 µg), doripenem (10 µg), tetracycline (30 µg), chloramphenicol (30 µg), cefuroxime (30 µg), and cephalexin (30 µg).
3.3. COVID-19 Detection
The laboratory diagnosis with the molecular and specific targets was done for all the susceptive patients who were admitted to different wards by the COVID-19 laboratory of the hospital. Although the gold standard for COVID-19 diagnosis has not been introduced yet, we considered real-time polymerase chain reaction (RT-PCR) in addition to the clinical demonstration of COVID-19 as an approximately accurate method (14). The data were analyzed with IBM SPSS software version 26.
3.4. Statistical Analysis
The data were analyzed using the software SPSS version 26, and the chi-square test was used for analyzing the data statically.
4. Results
4.1. Patients
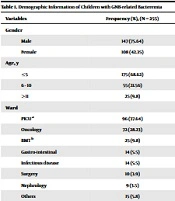
A total of 255 patients with GNB-related bacteremia were included in this study (including 57.64% (147/255) males and 42.35% (108/255) females). Approximately 68.6% (175/255) of the patients were under the age of 5 years, and 37.64% (96/255) of the patients were admitted to PICU. According to the real-time PCR results, COVID-19 infection was confirmed in 25% (64/255) of the patients, and the death rate was recorded in 25.5% (65.255) of children with GNB-related bacteremia (Table 1).
| Variables | Frequency (%), (N = 255) |
|---|---|
| Gender | |
| Male | 147 (75.64) |
| Female | 108 (42.35) |
| Age, y | |
| ≤5 | 175 (68.62) |
| 6 - 10 | 55 (21.56) |
| ≥11 | 25 (9.8) |
| Ward | |
| PICU a | 96 (37.64) |
| Oncology | 72 (28.23) |
| BMT b | 25 (9.8) |
| Gastro-intestinal | 14 (5.5) |
| Infectious disease | 14 (5.5) |
| Surgery | 10 (3.9) |
| Nephrology | 9 (3.5) |
| Others | 15 (5.8) |
| COVID-19 | |
| Yes | 64 (25) |
| No | 191 (75) |
| Death | 10 (9.8) |
| Yes | 65 (25.5) |
| No | 190 (74.5) |
a bone marrow transplantation
b pediatric intensive care unit
4.2. The Frequency of GNB Isolated from the Blood of Children
Among all the GNB isolated from blood samples of children with bacteremia, Enterobacteriaceae spp. with a frequency of 43.5% (111/255) were the most frequent isolates, and the most frequent Enterobacteriaceae members belonged to Klebsiella pneumonia 79.27% (88/111), Enterobacter spp. 10.8% (12/111), Escherichia coli 8.1% (9/111), Yersinia 0.9% (1/111), and Citrobacter 0.9% (1/111). The other GNB strains included Pseudomonas spp. 33.7% (86/255), Acinetobacter spp. 21.6% (55/255), and Stenotrophomonas spp. 1.2% (3/255).
4.3. The Resistant Profile of Gram-negative Bacteria and Frequency of Multi-drug Resistant Strains
The high resistance percentage was recorded against all of the antibiotics among GNB isolated from blood samples of children with bacteremia. K. pneumonia strains showed the most resistant profile against ampicillin and ampicillin-sulbactam, with 94.31%, and the other members of the Enterobacteriaceae family were resistant to most of the antibiotics. Acinetobacter spp. were resistant to most of the antibiotics, especially ampicillin. Pseudomonas spp. showed lower resistance levels to the antibiotics, but 77% of them were resistant to ceftazidime (Table. 2)
| Antibiotics | Escherichia spp. (N = 9) | Klebsiella spp. (N = 88) | Enterobacter spp. (N = 12) | Citrobacter spp. (N = 1) | Yersinia spp. (N = 1) | Acinetobacter spp. (N = 55) | Stenotrophomonas (N = 3) | Pseudomonasspp. (N = 86) |
|---|---|---|---|---|---|---|---|---|
| GM (10 µg) | 7 (77.77) | 80 (90.9) | 11 (91.6) | 0 | 0 | 36 (65.45) | 1 (33.33) | 48 (55.81) |
| TOB (10 µg) | 8 (88.88) | 81 (92.04) | 11 (91.6) | 1 (100) | 0 | 49 (89.09) | 3 (100) | 50 (58.13) |
| AN (30 µg) | 8 (88.88) | 71 (80.6) | 8 (66.66) | 1 (100) | 0 | 29 (52.72) | 3 (100) | 35 (40.69) |
| CIP (5 µg) | 5 (55.55) | 57 (62.5) | 7 (58.33) | 0 | 1 (100) | 14 (25.45) | 2 (66.66) | 21 (24.4) |
| LVX (5 µg) | 6 (66.66) | 68 (77.27) | 10 (83.33) | 1 (100) | 0 | 19 (34.54) | 3 (100) | 21 (24.4) |
| PIP (30 µg) | 6 (66.66) | 81 (92.04) | 11 (91.6) | 1 (100) | 1 (100) | 49 (89.9) | 3 (100) | 59 (68.6) |
| CAZ (30 µg) | 8 (88.88) | 81 (92.04) | 11 (91.6) | 1 (100) | 1 (100) | 50 (90.9) | 3 (100) | 67 (77.9) |
| PTAZ (110 µg) | 5 (55.55) | 80 (90.9) | 11 (91.6) | 1 (100) | 1 (100) | 49 (89.9) | 3 (100) | 58 (67.44) |
| CFM (30 µg) | 5 (55.55) | 80 (90.9) | 11 (91.6) | 1 (100) | 0 | 47 (85.45) | 3 (100) | 56 (65.11) |
| AZT (30 µg) | 5 (55.55) | 71 (80.6) | 11 (91.6) | 1 (100) | 0 | 48 (87.27) | 3 (100) | 55 (63.95) |
| IMP (10 µg) | 4 (44.44) | 66 (75) | 11 (91.6) | 1 (100) | 1 (100) | 46 (83.63) | 2 (66.66) | 57 (66.27) |
| MEM (10 µg) | 7 (77.77) | 80 (90.9) | 9 (75) | 0 | 1 (100) | 48 (87.27) | 3 (100) | 59 (68.6) |
| SXT (2.5 µg) | 8 (88.88) | 75 (85.22) | 11 (91.6) | 0 | 0 | 31 (56.36) | 2 (66.66) | _ |
| CTX (30 µg) | 9 (100) | 79 (89.77) | 11 (91.6) | 1 (100) | 0 | 49 (89.9) | 3 (100) | _ |
| AMP (10 µg) | 7 (77.77) | 83 (94.31) | 11 (91.6) | 1 (100) | 1 (100) | 50 (90.9) | 3 (100) | _ |
| APS (20 µg) | 9 (100) | 83 (94.31) | 10 (83.33) | 1 (100) | 1 (100) | 49 (89.9) | 3 (100) | _ |
| DOX (30 µg) | _ | _ | _ | _ | _ | 44 (80) | 3 (100) | _ |
| DOR (10 µg) | 9 (100) | 82 (93.18) | 11 (91.6) | 1 (100) | 1 (100) | 44 (80) | 3 (100) | _ |
| TET (30 µg) | 9 (100) | 79 (89.77) | 11 (91.6) | 1 (100) | 0 | 47 (85.45) | 3 (100) | _ |
| CN (30 µg) | 8 (88.88) | 73 (82.95) | 11 (91.6) | 1 (100) | 0 | 47 (85.45) | 3 (100) | _ |
| CXM (30 µg) | 9 (100) | 82 (93.18) | 11 (91.6) | 1 (100) | 1 (100) | 47 (85.45) | 3 (100) | _ |
| CFX (30 µg) | _ | _ | _ | _ | _ | 47 (85.45) | 3 (100) | _ |
Abbreviations: GM, gentamycin (10 µg); TOB, tobramycin (10 µg); AN, amikacin (30 µg); CIP, ciprofloxacin (5 µg); LVX, levofloxacin (5 µg); PIP, piperacillin (30 µg); CAZ, ceftazidime (30 µg); PTAZ, piperacillin-tazobactam (110 µg); CFM, cefepime (30 µg); AZT, aztreonam (30 µg); IMP, imipenem (10 µg); MEM, meropenem (10 µg); SXT, trimethoprim-sulfamethoxazole (2,5 µg); CTX, cefotaxime (30 µg); AMP, ampicillin (10 µg); APS, ampicillin-sulbactam (20 µg); DOX, doxycycline (30 µg); DOR, doripenem (10 µg); TET, tetracycline (30 µg); CN, chloramphenicol (30 µg); CXM, cefuroxime (30 µg); and CFX, cephalexin (30 µg).
a Values are expressed as No. (%).
4.4. MDR Strains and COVID-19 in Children
A multitude of different MDR patterns was detected in the resistant GNB isolated from blood samples. Overall, 86.66% (221/255) of the GNB were MDR, and the most prevalent pattern was being resistant to imipenem, meropenem, ceftazidime, piperacillin, piperacillin-tazobactam, cefepime, aztreonam, tobramycin, and gentamycin with an average percentage of 80.1%. The frequency of MDR strains among GNB was as follows: Enterobacteriaceae spp. 91.8% (102/111), Pseudomonas spp. 77.9% (67/86), Acinetobacter spp. 89% (49/55), and Stenotrophomonas spp. 100% (3/3). Out of 255 children with GNB-related bacteremia, 25.1% (64/255) of them had confirmed the COVID-19 test and 93.7% (60/64) of these patients had both MDR bacteremia and COVID-19. Moreover, the correlation between COVID-19 and MDR bacteremia was investigated according to Table 3, and there was a significant relationship between MDR bacteremia caused by GNB and COVID-19. The death rate of patients with both MDR bacteremia and COVID-19 was 43.33% (26/60) among these children.
| GNB Strains | Frequency of MDR in Blood Samples | COVID-19 Test of the Patients with MDR Bacteremia | P-Value |
|---|---|---|---|
| Pseudomonas spp. | 77.9% (67/86) | 19.4% (13/67) | 0.002 |
| Acinetobacter spp. | 89.09 (49/55) | 12.24% (6/49) | |
| K. pneumonia | 94.33% (83/88) | 36.14% (30/83) | |
| E.coli | 88.88% (8/9) | 62.5% (5/8) | |
| Enterobacter genus | 83.33% (10/12) | 40% (4/10) | |
| Yersinia spp. | 0 | 0 | |
| Citrobacter spp. | 100 (1/1) | 0 | |
| Stenotrophomonas spp. | 100% (3/3) | 66.66% (2/3) |
5. Discussion
There are miscellaneous potential risk factors that could complicate the process of bacteremia treatment. The latest challenge is COVID-19, which could affect patients’ lives and the spread of infection (15). One of the pivotal risk factors here is increasing the length of stay in the hospital, which could lead to boosting the prevalence of viral and bacterial superinfections. As patients who are admitted to the different wards of the hospital have been transported to the other wards with the change of their circumstances, such as BSIs or positive COVID-19 tests, there is a great chance of infection spreading throughout the hospital (16). At the time of writing this article, the correlation of bacteremia, particularly the ones caused by MDR strains and COVID-19, is not well understood in children. Clarifying the role of MDR bacteremia and COVID-19 in children and identification of carriers is an important point because they could be asymptomatic; while they do not show severe symptoms, they will spread resistant genes, and COVID-19 in both the hospital and the community (17).
Herein, we studied GNB isolated from blood samples of children with bacteremia who were admitted to the different wards of the hospital and their resistant profile. All of them were followed for COVID-19 in the case of susceptive clinical demonstrations such as fever and cough. We considered gender, age, and the wards where patients were hospitalized as three useful variables. Although there was not a significant relationship between the age and sex of the patients with MDR bacteremia and COVID-19 superinfection, the correlation between the ward of the admission and the co-infection was significant (P-value = 0.011), 32% of the patients with both MDR bacteremia and COVID-19 were admitted to the oncology unit, which is a high percentage versus the situation of the other patients who were admitted in the other units. Based on a study in 2021 in the USA, COVID-19 could affect hematologic malignancy patients more than other patients (18). However, based on the results of other studies, based on only COVID-19 mortality rates, most of the patients were admitted to the PICU (19).
The most frequent GNB isolated from blood samples of children with bacteremia was Enterobacteriaceae spp., with 43.5%, and K. pneumoniae was the most frequent genus of this family, with 79.27%. According to a study that was done in 2015 in Italy, the most frequent GNB isolated from children with bacteremia belonged to Enterobacteriaceae spp. with 53% (20).
The second and third frequent GNB isolated from the blood samples were 33.7% Pseudomonas spp. and 21.6% Acinetobacter spp. Previous studies have reported the high frequency of these GNB in Iranian hospitals in which 42.22% of the GNB belonged to Acinetobacter spp. (21). In the current study, 86.66% of all the GNB were MDR, and about 80% of them showed a wide range of resistance profiles against imipenem, meropenem, ceftazidime, piperacillin, piperacillin-tazobactam, cefepime, aztreonam, tobramycin, and gentamycin. The results of previous studies showed a lower level of resistance to these antibiotics (22). This difference emphasizes the importance of the effects of COVID-19 and how it affects the rise of resistance genes among GNB. The use of some antibiotics that are known as the last line of treatment, like carbapenems as prophylaxis regimens in patients with COVID-19, may help emerge new resistance mechanisms and spread the antimicrobial resistant genes. In addition, 25% of all the patients with bacteremia were positive for COVID-19. Other studies during the pandemic showed the overwhelming capacity of blood cultures in hospitals. Although the general rate of COVID-19 is not high among patients with bacteremia, the sharp trend of reports is concerning (23).
The main cause of bacteremia in hospitalized patients, particularly during the COVID-19 pandemic, was detected as nosocomial pathogens and healthcare-associated infections (HAIs). All of the current situations in the hospitals, including the length of stay, as well as the use of broad-spectrum antibiotics, ventilators, and catheters, are optimal for spreading MDR strains throughout the hospital and colonizing both patients and the staff (24). In this study, 93.7% of the patients with confirmed COVID-19 tests had MDR bacteremia. Diagnosis of co-infections is uncommon and could be a challenge in children (25). The death rate was 43.33% among children with MDR bacteremia and COVID-19. To the date of writing this article and based on our knowledge, an authentic paper was not published to compare our results with it. Our data highlighted the high frequency of MDR strains isolated from blood samples and the high prevalence of COVID-19 among children with MDR bacteremia. The high mortality rate remarked the necessity of designing a new diagnostic approach for characterizing and detecting common etiologies of bacteremia to prevent infection and decrease mortality risk factors in hospitalized children. There is a plethora of solutions for source tracking and preventing the MDR strains from spreading. In this regard, molecular epidemiology and molecular typing studies could be introduced as the best ones.
5.1. Conclusions
The high frequency of MDR-GNB-related bacteremia in children was reported in this study. The results of this study highlighted the critical role of COVID-19 in children with MDR-GNB-related bacteremia. The high frequency of bacteremia caused by MDR-Enterobacteriaceae spp. is concerning in the studied population. Implementation of infection prevention and control programs is necessary to reduce concomitant infections and mortality rates in hospitalized children.