1. Background
Pseudomonas aeruginosa is one of the main nosocomial pathogens, which can cause mortality rates of 30 - 50% in patients with bacteremia and even 70% in patients with nosocomial pneumonia. This pathogen can survive in hospital settings, food, disinfectants, clinical specimens, and aquatic and soil habitats due to high adaptability and grabbing antibiotic resistance (1, 2). The resistance among clinical isolates has been classified into three categories, including multidrug-resistant (MDR), extremely drug-resistant (XDR), and pan-drug-resistant (PDR), which are serious causes of healthcare-associated infections worldwide.
After the emergence of carbapenem-resistant isolates, polymyxins were frequently used for Pseudomonas infections therapy. After a while, polymyxin-resistant strains emerged among hospitalized patients. Consequently, P. aeruginosa became the main threat to hospitalized patients (3). Several molecular typing schemes have been described for epidemiological studies and to study genetic linkages of bacteria, especially in nosocomial infections (4). Among these methods are biotyping, phage typing, serotyping, and genotyping, which are used based on purposes, conditions, and facilities of healthcare laboratories (5). The enterobacterial repetitive intergenic consensus-polymerase chain reaction (ERIC-PCR) method has been known for its low complication, rapid, reproducible, and precise results. Also, it is cost-benefit compared with other typing methods (6).
2. Objectives
This study aimed to investigate resistance patterns and genotypes of P. aeruginosa isolated from patients with nosocomial infections using the ERIC-PCR method.
3. Methods
3.1. Bacterial Strains
In this experimental study, 149 samples were collected from September 2020 to February 2021 from patients hospitalized in a teaching hospital in Tehran, Iran. The clinical samples included urine, throat, CSF, blood, sputum, ascites, pleural fluid, and wound samples collected from patients hospitalized in different wards.
3.2. Isolation and Identification of Pseudomonas aeruginosa
Each specimen was subjected to a particular isolation method. Of 149 samples, 76 were detected as P. aeruginosa based on initial identification by biochemical testing, including Gram staining, oxidase, citrate, oxidative-fermentative, TSI, SIM, urease, catalase, and growth at 42°C.
3.3. Antibiotic Susceptibility Testing
The antibiotic susceptibility analysis was performed by the disc diffusion method according to the CLSI guideline 2020 (7) using antibiotic disks (Mast Group Ltd, Merseyside, UK). According to the CLSI guideline, eight antibiotic categories would be tested to determine MDR, XDR, and PDR strains. They were aminoglycosides, carbapenems, cephalosporins, fluoroquinolones, penicillins/ß-lactamase inhibitors, monobactams, phosphonic acids, and polymyxins. Moreover, the minimum inhibitory concentration (MIC) for colistin and polymyxin B were measured using the micro-broth dilution method according to the CLSI (7). Antibiotics powders were purchased from Sigma-Aldrich Inc. Pseudomonas aeruginosa ATCC 27853 was used as a reference in all microbial and molecular tests.
3.4. DNA Extraction and Molecular Confirmation Test
The whole genome of all initially identified P. aeruginosa isolates were extracted using Qiagen whole genome extraction kit according to the manufacturer's instruction (Qiagen, Germany). The quality of extracted DNA was checked by measuring the absorbance ratio at 260 and 280 nm with a spectrophotometer. Then, it was used as a DNA template in the Polymerase Chain Reaction (PCR) assay. The PCR was performed using specific primers for the peptidoglycan-associated lipoprotein (oprL) gene (forward 5'-ATGGAAATGCTGAAATTCGGC-3'and reverse 5'- CTTCTTCAGCTCGACGCGACG-3'), which particularly determines P. aeruginosa strains (8). The PCR mixture was prepared in 25 µL reactions consisting of 12.5 µL ready to use master mix (including MgCl2, dNTPs, and Taq polymerase), 0.25 µL of 10 nmol.µL-1 of each primer, 5 µL of extracted DNA, and sterile nuclease-free water up to 25 µL final volumes. The PCR program was started with initial denaturation (93°C for 5 min), followed by 30 cycles, including denaturation (93°C for 1 min), annealing (57°C for 1 min), and extension (72°C for 1 min); final extension was set at 72°C for 5 min. The PCR products were subjected to horizontal electrophoresis on a 1.5% agarose gel using a 1 kb DNA ladder and visualized with commercial DNA safe stain (SinaClon BioScience Co. Iran). Images were captured using the UVItec gel documentation system (Cleaver Scientific Ltd, United Kingdom).
3.5. Enterobacterial Repetitive Intergenic Consensus-Polymerase Chain Reaction Genotyping
The whole genome of bacteria was used as a DNA template for the ERIC-PCR assay. The primer sets ERIC-1R, 5'ATGTAAGCTCCTGGGGATTCA3' and ERIC-2, 5'AAGTAAGTGACTGGGGTGAGCG3' were used to amplify the regions in the bacterial genome (9). The amplification was conducted using an Analytic Jena (Jena, Germany) thermal cycler with the following program: 95°C for 5 min, followed by 25 cycles of 94°C for 45 s, 55°C for 45 s, and 72°C for 1 min, with a final extension at 72°C for 5 min. The PCR mixture was prepared in a 25 µL final volume, containing 5 µL of the extracted DNA, 12.5 µL of PCR Master Mix, 0.5 µL of 10 pM of each primer, and 6.5 µL of distilled water. The PCR products were separated with gel electrophoresis on a 2% agarose gel using a 50 bp DNA ladder (Fermentas) and visualized through ultraviolet illumination.
3.6. Data Analysis
The statistical analysis was done using IMB SPSS version 26 software. The ERIC-PCR data were analyzed with Gel Compare II software version 6.6.11 (Applied Maths, Sint-Martens-Latem, Belgium). The isolates were clustered on the Dice similarity coefficient on the basis of the Unweighted Pair Group Method with Arithmetic Mean (UPGMA) analysis.
4. Results
4.1. Clinical Samples and Bacterial Strains
A total of 149 clinical samples were collected from hospitalized patients admitted to a teaching hospital affiliated with Iran University of Medical Sciences, Tehran, Iran, for six months. The clinical samples were various, mostly throat samples (94/149). The patients had been admitted to various hospital wards, mostly the internal ICU (79/149). Seventy-six out of 149 (51%) clinical specimens were positive for P. aeruginosa, initially identified by biochemical tests and confirmed by harboring the 504 bp DNA fragment belonging to the oprL gene (Figure 1). The P. aeruginosa isolates were detected with the highest frequency in throat samples (52.6%). In comparison, ascites fluid samples showed the lowest frequency (2.6%). The hospitalized elderlies (> 65 years old) were at a higher risk for acquiring P. aeruginosa infection, such that 43 out of 76 (56.6%) strains were isolated from these patients. Most P. aeruginosa isolates were from patients hospitalized in different ICUs (85.3%). On the contrary, the other wards showed a low spread of P. aeruginosa infection (2.7% for each).
4.2. Antimicrobial Resistance Pattern and MDR, XDR, and PDR Strains Determination
All isolates were subjected to susceptibility testing against 17 antibiotics according to the CLSI guidelines for determining MDR, XDR, and PDR strains. The highest resistance was observed against aminoglycosides, carbapenems, cephalosporins, and fluoroquinolones (almost more than 70%). At the same time, polymyxins, especially polymyxin B, had the lowest resistance rate (9.2%). The susceptibility details are shown in Table 1. The isolates showed high diversity in their antibiotic resistance patterns, and 41 different antibiotypes were recognized. Based on the CLSI description, MDR was defined as resistant to at least one agent in three or more antimicrobial categories; XDR was defined as non-susceptible to at least one agent in all, but two or fewer antimicrobial categories," and PDR was defined as non-susceptible to all agents in all antimicrobial categories. Based on these criteria, 66 (86.8%) strains were determined as MDR, 62 (81.5%) as XDR, and 10 (13.2%) as non-MDR. Four (5.3%) strains resisted all tested antibiotics and were classified as PDR.
| Antimicrobial Categories and Agents | Susceptible | Intermediate | Resistant |
|---|---|---|---|
| Aminoglycosides | |||
| Gentamicin | 18 (23.7) | 3 (3.9) | 55 (72.4) |
| Tobramycin | 18 (23.7) | 0 (0) | 56 (76.3) |
| Amikacin | 25 (32.9) | 5 (6.6) | 46 (60.5) |
| Netilmicin | 18 (23.7) | 2 (2.6) | 56 (73.7) |
| Carbapenems | |||
| Imipenem | 15 (19.7) | 8 (10.5) | 53 (69.7) |
| Meropenem | 15 (19.7) | 10 (13.2) | 51 (69.7) |
| Doripenem | 11 (14.5) | 2 (2.6) | 63 (82.9) |
| Cephalosporins | |||
| Ceftazidime | 19 (25) | 0 | 57 (75) |
| Cefepime | 17 (22.4) | 2 (2.6) | 57 (75) |
| Fluoroquinolones | |||
| Ciprofloxacin | 15 (19.7) | 2 (2.6) | 59 (77.6) |
| Levofloxacin | 16 (21.1) | 4 (5.3) | 56 (73.7) |
| Penicillins/ß-lactamase inhibitors | |||
| Ticarcillin-clavulanic acid | 11 (14.5) | 16 (21.1) | 49 (64.5) |
| Piperacillin-tazobactam | 36 (47.4) | 8 (10.5) | 32 (42.1) |
| Monobactams | |||
| Aztreonam | 24 (31.6) | 3 (4.9) | 49 (64.5) |
| Polymyxins | |||
| Polymyxin B | 69 (90.8) | 0 | 7 (9.2) |
| Colistin | 49 (64.5) | 0 | 27 (35.5) |
| Fosfomycins | |||
| Fosfomycin | 10 (13.2) | 0 | 66 (86.8) |
a Values are expressed as No. (%).
4.3. Genotyping of Isolates
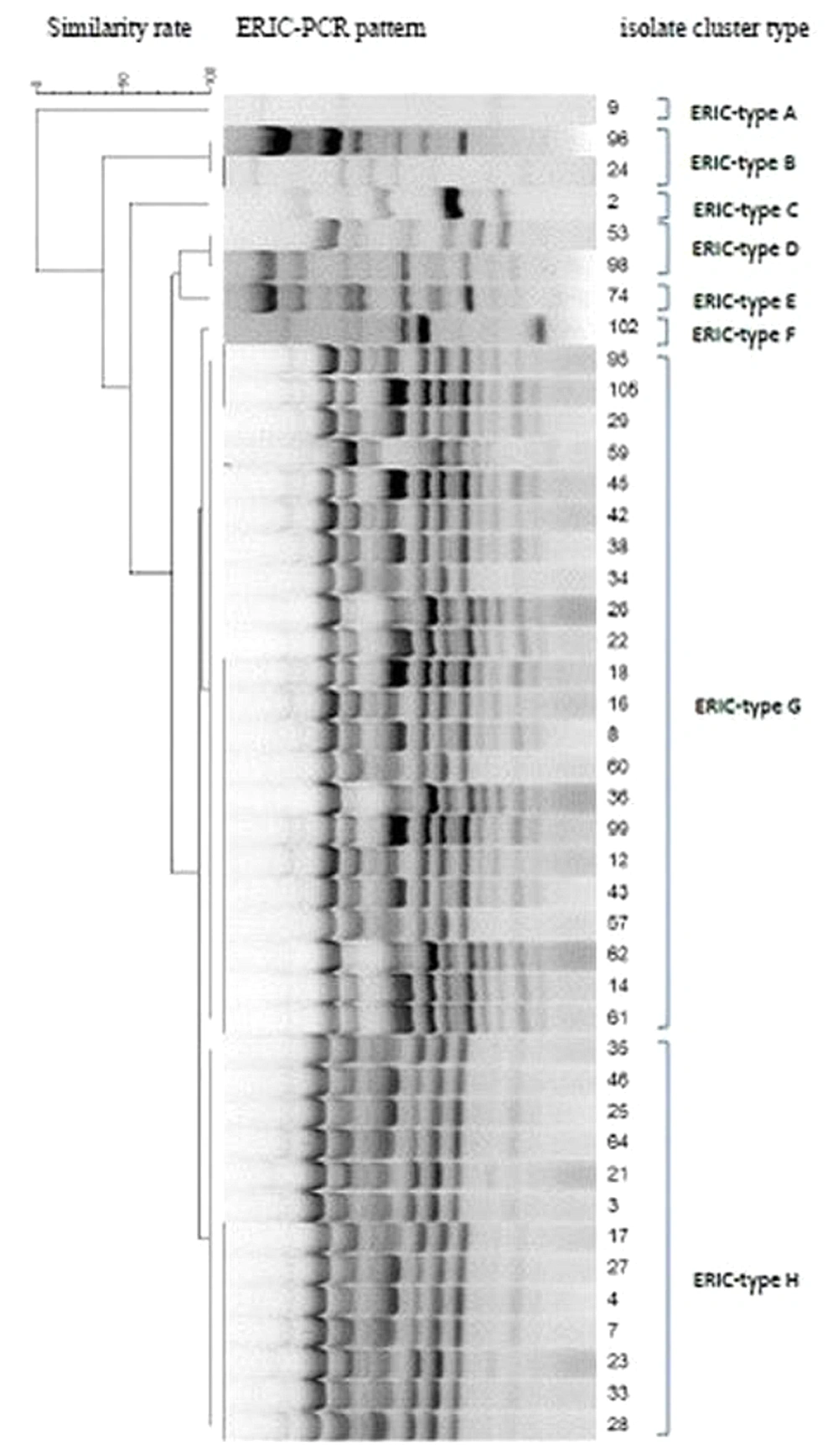
The ERIC-PCR gel analysis exhibited eight ERIC types (E-type) with 4-17 bands ranging from 50 to 2000 bp (Figure 2). The predominant ERIC-PCR type (E-type) was related to G with 22 (28.9%) isolates, followed by E-type H with 13 (17.1%) isolates. Each of the A, C, E, and F types had a unique genetic diversity and only included one isolate. E-type A belonged to the sputum sample, while E-types C, E, and F belonged to the throat samples. The E-type D consisted of two isolates that belonged to different clinical samples. The E-type G included a wide range of clinical samples and almost consisted of isolates from all types of specimens; on the contrary, the E-type H was related to urine and throat samples. The statistical analysis showed no significant relationship between the E-types and sample types. On the contrary, ward-wise analysis of E-types showed some correlations with the isolates. For instance, the isolates belonging to E-type B were related to the general ICU, and isolates of the E-types H and G belonged to the internal ICU. Notably, all PDR strains were from the internal ICU, three of which belonged to E-type G, and the remaining one was of E-type H, the second dominant E-type in the internal ICU. There was no significant correlation between E-types and antibiotic resistance patterns. However, all strains of one E-type showed resistance to some antibiotics (Table 2).
| E-Types | Antibiotic Resistance Pattern | Strains, No. (%) |
|---|---|---|
| A | GEN-TOB-AMI-NET-IPM-MEM-DOR-CAZ-CPM-CIP-LVX-TIM-PTZ-FOS | 1 (1.3) |
| B | GEN-TIM-FOS | 2 (2.6) |
| C | GEN-TOB-AMI-NET-IPM-MEM-DOR-CIP-LVX-PTZ-ATM-FOS | 1 (1.3) |
| D | GEN-TOB-AMI-NET-IPM-DOR-CAZ-CPM-CIP-LVX-TIM-ATM | 2 (2.6) |
| E | GEN-TOB-AMI-NET-IPM-DOR-CAZ-CPM-CIP-LVX-ATM-FOS | 1 (1.3) |
| F | GEN-TOB-AMI-NET-IPM-MEM-DOR-CAZ-CPM-TIM-PTZ-ATM-PB-FOS-CL | 1 (1.3) |
| G | TIM-FOS | 22 (28.9) |
| H | DOR-TIM-FOS | 13 (17.1) |
| Total | 76 |
Abbreviations: GEN, gentamicin; TOB, tobramycin; -AMI, amikacin; NET, netilmicin; IPM, imipenem; MEM, meropenem; DOR, doripenem; CAZ, ceftazidime; CPM, cefepime; CIP, ciprofloxacin; LVX, levofloxacin; TIM, ticarcillin-clavulanic acid; PTZ, piperacillin-tazobactam; ATM, aztreonam; FOS, fosfomycin; CL, colistin; PB, polymyxin B.
5. Discussion
Our study observed 41 antibiotic resistance patterns and eight E-types among P. aeruginosa isolates. This study observed the highest P. aeruginosa prevalence among ICU patients; most were isolated from throat samples. One reason for these findings can be related to the high sickness of patients hospitalized in ICUs with weak immune systems. Moreover, abuse or overuse of antibiotics and medical instruments in ICUs intensifies the circumstances. Other studies in Iran also proved that the emergence of P. aeruginosa is significantly higher in ICUs than in other hospital units (10, 11). The most probable specimens contaminated with P. aeruginosa in our study were throat, followed by urine samples. In the study by Vaez et al., however, the highest frequency of P. aeruginosa was related to ICU patients, and urine samples were primarily infected with P. aeruginosa (11). In another study by Izadi Pour Jahromi et al., burn patients and pediatrics were reported as high-risk groups for Pseudomonas infections, and isolates showed notable and drastic resistance to various antibiotics (12). Some other countries reported that the most frequent Pseudomonas infections occurred in respiratory tracts, most of which were among ICU patients (13-15).
Our isolates showed high resistance to aminoglycosides, carbapenems, cephalosporins, and fluoroquinolones. The most effective antibiotics belonged to polymyxins, especially polymyxin B. The MDR and XDR were detected in 86.8% and 81.5% of our isolates, respectively, which proves the high antibiotic resistance rate among P. aeruginosa strains. Although different rates have been reported in Iran and other countries for MDR and XDR P. aeruginosa, all agree with the upward trend of resistant strains emergence, especially XDR strains (16-18). In our study, four strains were classified as PDR, showing the increasing emergence of PDR strains. The emergence of PDR strains was not reported in previous studies, or at least it was very low (13, 15, 16). In contrast, in our study, the detection of four PDR strains out of 76 isolates proves that we are near the end of the antibiotic era.
Studies of genotyping and molecular epidemiology of P. aeruginosa strains showed that the genetic diversity is catastrophically high among clinically resistant P. aeruginosa strains than in susceptible or environmental strains (19). The molecular epidemiologic studies warned about the increasing emergence and spread of P. aeruginosa high-risk clones worldwide, a major concern in preventing and controlling nosocomial infections (20). Thus, the source tracking and molecular epidemiologic information of resistant P. aeruginosa play a key role in infection treatment and control.
The present study observed eight E-types among resistant P. aeruginosa named from A to H alphabetic letters. The dominant E-types among our isolates belonged to G and H, composed of strains isolated from ICU patients. Although a high diversity in resistance patterns was observed among strains belonging to these two E-types, all were resistant to ticarcillin-clavulanic acid and fosfomycin and showed almost 90% similarities in their ERIC patterns. The isolates related to these two E-types can be considered dominant resistant strains spread in the internal ICU. The E-type A was unique among isolates with no similarity with other E-types from a patient hospitalized in the surgical ICU. Moreover, E-types C, E, and F were each composed of only one strain isolated from throat samples and showed a 50% similarity with each other. Notably, although the former E-types showed only 50% similarity, they showed resistance to some antibiotics in common. According to the analysis, no correlations were observed between E-types and sample types or antibiotic resistance patterns (P ≥ 0.05), while the strains with the same E-types were almost related to the same ward in the hospital. In the study by other researchers from Iran, the ERIC-PCR results of P. aeruginosa isolates could not be useful for predicting antibiotic resistance patterns because the strains with the same E-types could possess different antibiotic resistance profiles (21).
On the one hand, the current study showed several orphan clusters indicating the high rate of genetic diversity among isolates; on the other hand, we found a dominant cluster with two different E-types with considerable isolate numbers, all colonized and isolated from patients in the internal ICU. The diversity among E-types for P. aeruginosa is not rare, and most of the studies worldwide reported some sort of diversity (22-27). The point is that among these various genotypes, some can dominate in a single ward and spread to other hospital sites, making it much more difficult to eradicate from hospitals. The main threat here is regarding the PDR strains since all were isolated from the same ward and belonged to the same genetic cluster. They can be considered nosocomial pathogens and should be deliberated as a critical threat in an emerging outbreak in a hospital.
5.1. Conclusions
We observed a heterogeneous population of dominant P. aeruginosa strains in different hospital wards. However, they possessed different resistance patterns, making infection control and patient treatments more difficult and time-consuming for the healthcare system. In order to disclose and eradicate the resistant P. aeruginosa strains, the genetic and antibiotic profiling of any dominant local colon must be considered a critical strategy and guideline for the hospital infection control committee.