1. Context
Antimicrobial resistance (AMR) is defined as the resistance of a microorganism to an antibiotic that was previously effective in treating the infections caused by it. Based on the World Health Organization (WHO) report in 2014, AMR is becoming a worldwide public health threat that can deteriorate the clinical benefits of current important antimicrobial agents. The WHO indicates seven multi-resistant bacteria as the major global concern of AMR. According to the report, Iran is located in a geographical region with high resistance to more than five of these multi-resistance bacteria (1). One of the most substantial objectives of tackling AMR is to strengthen knowledge and improve the evidence through surveillance and research activities. Therefore, the existence of an antimicrobial surveillance system is required to estimate the burden of the AMR phenomenon at both national and international levels (2).
As an effective tool, the scoping review is used as a foundation for the formulation of an action plan and a research agenda on a specific broad topic (3). This kind of review can clarify the current research situation to identify the existence of sufficient evidence to make clear and well-advised recommendations. A scoping review can also map the existing literature regarding nature, features, and volume. Furthermore, a scoping review explains a topic’s conceptual borders and the requirement for further knowledge synthesis and reflects the gaps existing in publications (4, 5).
In recent years, a growing number of scoping reviews have been published worldwide for various purposes, mainly on health topics (3). In Iran, some scoping reviews have been conducted on medical subjects, such as chronic diseases or tobacco control; however, none of them has addressed AMR (6, 7). A national survey on healthcare-associated infections in Iran showed that the pattern and severity of AMR in different provinces are different. However, in general, urinary tract infections and respiratory infections, in addition to Escherichia coli, Acinetobacter, and Klebsiella, accounted for the largest proportion of healthcare-associated infections, diseases, and pathogens in Iran, respectively (8).
To meet the future challenges of infectious diseases and minimize the consequences of growing AMR in Iran, a better understanding of relevant studies is required; therefore, future research can be highly prioritized and supported accordingly (9, 10). This scoping review was carried out to provide an evidential base for identifying priority research and the characteristics of the published reports on AMR in Iran to identify potential gaps.
2. Methods
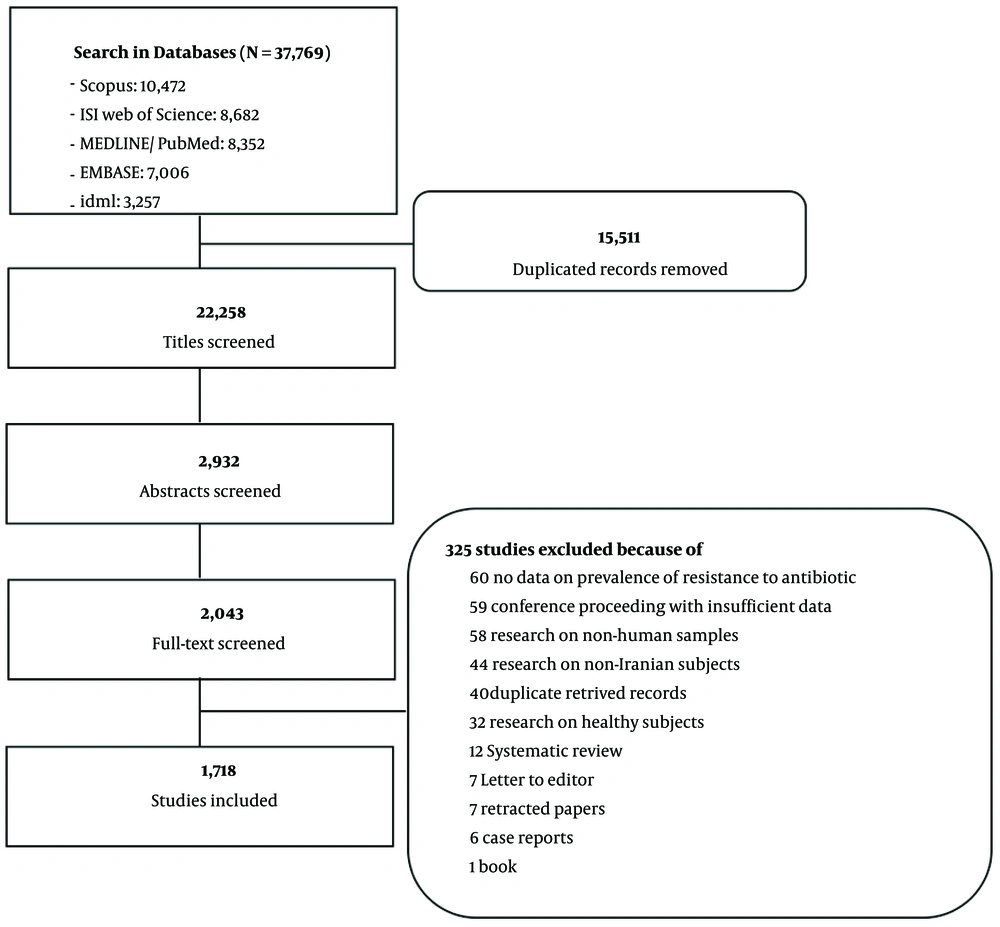
A sensitive systematic search was conducted to identify studies on antibiotic resistance clinical research in Iran in Scopus (since 1970), Institute for Scientific Information (ISI), Web of Science (since 1953), MEDLINE/PubMed (since 1966), EMBASE (since 1966), and Iranian Database of Medical Literature (IDML, http://idml.research.ac.ir; since 2008) in February 2018. The search study and screening are depicted in the Supplementary File 1.
This review included primary studies that evaluated the resistance or susceptibility of antibiotics to any bacteria in an Iranian population without language restrictions. Studies on non-human or non-Iranian samples, a healthy population, and with no reporting data on antibiotic resistance were excluded. Review articles, case reports, letters to the editor, conference proceedings with insufficient data on AMR prevalence, and papers that have been retracted were also excluded.
Four reviewers (AB, FT, EK, and MN) extracted the data, and three reviewers (BM, AB, and FS) verified the extracted data. The authors extracted the data on the characteristics of the scientific report (type of study, first author, publication year, and journal name and its publisher in terms of local or international) and the study (start date, duration, city, province, sample size, age of patients, type of infection, type of bacteria, and the list of antibiotics tested for resistance or susceptibility in each study).
The third and fourth levels of anatomical therapeutic chemical (ATC) classification were used to classify the antibiotics. This review assessed the type of infections based on 17 categories: Anaerobic infection, antibiotic-associated diarrhea, atypical pneumonia, foodborne disease, gastrointestinal disease, hospital infections, meningitis, respiratory infections, sexually transmitted diseases, tuberculosis, a wide range of clinical infections, zoonotic infection, and common and uncommon non-specific bacteria. The number of studies based on the province was analyzed since the prevalence of AMR might differ based on the location. R package software version 3.6.0 (2019-04-26) (R Foundation for Statistical Computing, Vienna, Austria; http://www.R-project.org) was used to plot the distribution of conducting sites of the studies based on Iranian provinces.
3. Results
3.1. Search and Selection of Studies
In total, 37,769 records were identified for potential inclusion, 1,718 of which, with 3,153,494 populations, fulfilled all inclusion criteria (the list of citations is available in Appendix 1 of the Supplementary File 1). The study selection process and the reasons for exclusion are outlined in the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) diagram (Figure 1).
The selected studies evaluated the susceptibility of 131 antibiotics (Table 1) to 82 types of bacteria (Table 2) by conducting 3,509 antibiotic resistance tests. Ofloxacin, ciprofloxacin, and gentamicin had the highest number of studies (1,148, 1,133, and 1,041, respectively), the highest number of samples (208,779, 207,513, and 205,272, respectively), and the highest number of tested bacteria (77, 76, and 75, respectively).
| Class of Antibiotic | Antibiotics (Number of Bacteria, Samples, and Studies) |
|---|---|
| Aminoglycosides | Amikacin (71, 157310, 783); Gentamicin (75, 205272, 1041); Kanamycin (45, 31960, 143); Neomycin (17, 1560, 16); Netilmicin (15, 5978, 34); Sisomicin (1, 72, 1); Streptomycin (32, 20191, 144); Tobramycin (39, 49396, 257) |
| Amphenicols | Chloramphenicol (58, 66148, 330); Florfenicol a (5, 308, 2) |
| Anti-infective and antiseptics | Fidaxomicin (1, 35, 1); Furazolidone (8, 2776, 24) |
| Anti-tuberculosis drugs and their combinations | Capreomycin (1, 672, 5); Cycloserine (1, 354, 4); Ethambutol (2, 6899, 38); Ethionamide (2, 824, 9); Isoniazid (2, 8933, 45); Para-aminosalicylic acid (1, 354, 4); Prothionamide (1, 78, 1); Pyrazinamide (2, 2126, 6); Rifabutin (2, 188, 2); Rifampicin (46, 39596, 235); Rifampicin-Colistin (1, 84, 1); Rifampicin-Tigecycline (1, 84, 1) |
| Beta-lactam antibacterials, penicillins, and their combinations | Amoxicillin (50, 29822, 174); Amoxicillin-Clavulanic acid (41, 19310, 152); Ampicillin (70, 119786, 517); Ampicillin-Sulbactam (23, 10110, 87); Azlocillin (1, 98, 2); Carbenicillin (26, 26933, 85); Clavulanic acid (9, 55, 2); Cloxacillin (37, 10955, 44); Dicloxacillin (2, 112, 2); Mecillinam (1, 48, 1); Methicillin (24, 8430, 60); Mezlocillin (3, 786, 9); Nafcillin (3, 560, 1); Oxacillin (49, 47596, 237); Penicillin (54, 61382, 280); Penicillin G (32, 7691, 38); Piperacillin (33, 32017, 224); Piperacillin-Tazobactam (37, 40104, 246); Tazobactam (10, 3530, 6); Tazobactam-Clavulanate (1, 103, 1); Ticarcillin (26, 11757, 91); Ticarcillin-Clavulanate (18, 4727, 33) |
| Carbapenems, monobactams, and other cephalosporins | Aztreonam (25, 34988, 255); Ceftaroline (3, 426, 2); Doripenem (4, 3967, 22); Ertapenem (11, 7076, 34); Imipenem (66, 115457, 725); Meropenem (41, 43897, 320) |
| Cephalosporins (first generation) | Cefalotin (52, 49215, 188); Cefazolin (47, 33817, 133); Cephalexin (55, 34132, 144); Cephalothin (7, 659, 3); Cephapirin (1, 40, 1); Cephradine (9, 1134, 4) |
| Cephalosporins (second generation) | Cefamandole (3, 364, 2); Cefotetan (16, 1532, 7); Cefoxitin (37, 25590, 164); Cefuroxime (35, 5053, 27) |
| Cephalosporins (third generation) and their combinations | Cefdinir (1, 123, 1); Cefditoren (3, 259, 2); Cefixime (56, 38592, 204); Cefoperazone (18, 1499, 11); Cefoperazone-Sulbactam (3, 246, 3); Cefotaxime (63, 95977, 560); Cefotaxime-Clavulanate (12, 3353, 21); Cefpodoxime (12, 7975, 54); Cefpodoxime- Clavulanate (2, 250, 2); Ceftazidime (65, 120641, 738); Ceftazidime-Clavulanate (12, 3134, 22); Ceftazidime-Vancomycin-Ampicillin (7, 293, 3); Ceftiofur a (5, 233, 2); Ceftizoxime (49, 53049, 171); Ceftriaxone (68, 107551, 552); Ceftriaxone-Tazobactum (10, 846, 2); Ceftriaxone-Vancomycin (7, 82, 2); Ceftriaxone-Vancomycin-Ampicillin (6, 428, 3) |
| Cephalosporins (fourth generation) and their combinations | Cefepime (51, 54353, 376); Cefepime-Clavulanate (1, 56, 1); Cefepime-Vancomycin (6, 18, 1); Cefpirome (6, 774, 2) |
| Macrolides, lincosamides, streptogramins, and their combinations | Azithromycin (42, 11013, 75); Clarithromycin (22, 7473, 64); Clindamycin (52, 62236, 285); Erythromycin (58, 77829, 413); Josamycin (1, 32, 1); Lincomycin (12, 2387, 12); Lincomycin-Spectinomycin (1, 245, 2); Metronidazole (20, 4670, 49); Pristinamycin (2, 114, 3); Quinolone (1, 54, 1); Quinupristin (1, 64, 1); Quinupristine-Dalfopristine (14, 10904, 59); Tylosin a (1, 11, 1) |
| Quinolone antibacterial | Ciprofloxacin (76, 207513, 1133); Enrofloxacin a (5, 1175, 9); Gatifloxacin (10, 3453, 26); Levofloxacin (29, 16053, 135); Lomefloxacin (5, 424, 3); Moxifloxacin (13, 1078, 11); Nalidixic acid (40, 81637, 350); Norfloxacin (43, 20797, 119); Ofloxacin (77, 208779, 1148); Pefloxacin (2, 72, 2) |
| Other antibacterial and their combinations | Bacitracin (1, 378, 1); Colistin (30, 18148, 168); Daptomycin (2, 277, 2); Fosfomycin (15, 2336, 18); Fosfomycin-Trometamol (1, 83, 1); Fusidic acid (10, 3855, 19); Linezolid (37, 23884, 153); Mupirocin (5, 4309, 37); Nitrofurantoin (51, 74676, 288); Novobiocin a (12, 393, 8); Polymyxin B (13, 9046, 63); Spectinomycin (10, 832, 8); Teicoplanin (16, 15249, 94); Vancomycin (56, 80830, 391) |
| Sulfonamides and trimethoprim | Sulfamethoxazole (29, 5323, 19); Trimethoprim (29, 6276, 31); Trimethoprim-Sulfamethoxazole (68, 170579, 791) |
| Tetracyclines and their combinations | Chlortetracycline (11, 2275, 2); Doxycycline (39, 10381, 81); Minocycline (14, 7559, 49); Oxytetracycline (3, 699, 8); Tetracycline (62, 103622, 615); Tigecycline (18, 10198, 79); Tigecycline-Colistin (1, 84, 1) |
Frequency of Reported Antibiotic Resistance or Susceptibility in Iranian Studies Based on the Type of Antibiotic
Table 2 shows the number of tests on antibiotic resistance or susceptibility for each bacteria, in addition to the sample size and the number of studies. Escherichia coli, Staphylococcus aureus, and K. pneumonia had the highest number of antibiotics tested for resistance (98, 90, and 82, respectively). The top three studied bacteria in terms of the sample size and the number of studies were E. coli (78,100; 470), S. aureus (45,730; 404), and Pseudomonas aeruginosa (40,991; 321). The AMR pattern of E. coli was the most frequently studied. Escherichia coli resistance to ofloxacin was determined in 59,872 isolates (353 studies), gentamicin in 59,771 isolates (345 studies), and ciprofloxacin in 58,861 isolates (347 studies). The detail of the aforementioned analysis is presented in the Supplementary File 2.
| Variables | Bacteria (Number of Antibiotics, Samples, and Studies) |
|---|---|
| Anaerobic bacteria | Bacteroides fragilis (27, 184, 3); Porphyromonas gingivalis (9, 120, 1); Propionibacterium acnes (16, 90, 2) |
| Atypical pneumonia | Bordetella pertussis (3, 790, 2); Legionella pneumophila (19, 172, 2); Mycoplasma spp. (11, 32, 1) |
| Bacteria-associated diarrhea | Clostridium difficile (23, 154, 4) |
| Bacteria that cause a wide range of clinical infections | Beta-hemolytic Streptococcus (33, 989, 7); Nontuberculous mycobacteria (21, 295, 7); Staphylococcus aureus (90, 45730, 404); Streptococcus agalactiae (38, 1944, 25) |
| Community-acquired pneumonia | Streptococcus pneumoniae (44, 3376, 63) |
| Endocarditis | Nonhemolytic streptococci (46, 124, 9) |
| Foodborne disease bacteria | Bacillus cereus (12, 3, 1); Brucella spp. (37, 436, 10); Campylobacter spp. (31, 236, 12); Clostridium perfringens (19, 85, 2); Listeria monocytogenes (25, 144, 5); Salmonella enterica (46, 1178, 17); Salmonella spp. (53, 2445, 42); Salmonella typhi (41, 497, 13); Salmonella typhimurium (24, 486, 8); Vibrio cholerae (22, 1953, 20) |
| Gastrointestinal disease bacteria | Helicobacter pylori (15, 5174, 58); Shigella dysenteriae (36, 493, 26); Shigella flexneri (42, 2160, 38); Shigella sonnei (45, 2786, 42); Shigella spp. (48, 2757, 43) |
| Hospital infection bacteria | Acinetobacter baumannii (76, 16393, 190); Coagulase-negative staphylococci (75, 16972, 200); Enterobacter spp. (74, 7287, 168); Enterococcus faecalis (56, 7428, 72); Enterococcus faecium (44, 3858, 63); Escherichia coli (98, 78100, 470); Klebsiella pneumoniae (89, 17028, 213); Pseudomonas aeruginosa (82, 40991, 321); Stenotrophomonas maltophilia (50, 9991, 12) |
| Melioidosis | Burkholderia spp. (14, 1, 1) |
| Meningitis bacteria | Haemophilus influenza (41, 478, 16); Neisseria meningitidis (33, 33, 6) |
| Respiratory infections bacteria | Alpha-hemolytic Streptococcus (34, 121, 6); Moraxella catarrhalis (9, 12, 2); Streptococcus pyogenes (42, 1997, 11); Streptococcus viridans (36, 381, 13) |
| Tuberculosis bacteria | Mycobacterium tuberculosis (35, 11170, 57) |
| Sexually transmitted diseases | Neisseria gonorrhoeae (20, 229, 5) |
| Common non-specific bacteria | Acinetobacter spp. (68, 3770, 99); Citrobacter spp. (67, 797, 77); Enterococcus spp. (72, 5492, 125); Klebsiella spp. (71, 7809, 150); Proteus spp. (68, 1349, 108); Pseudomonas spp. (61, 2780, 77); Serratia spp. (52, 528, 32); Staphylococcus spp. (52, 1708, 35); Streptococcus spp. (60, 585, 39) |
| Uncommon non-specific bacteria | Aerobacter spp. (18, 3, 1); Aeromonas spp. (14, 12, 1); Alcaligenes spp. (34, 30, 6); Bacillus spp. (32, 121, 7); Bacteroides spp. (18, 30, 1); Branhamella spp. (15, 56, 2); Capnocytophaga spp. (16, 1, 1); Cedecea spp. (15, 7, 1); Clostridium spp. (18, 8, 1); Corynebacterium spp. (28, 38, 5); Edwardsiella spp. (20, 3, 3); Escherichia spp. (10, 65, 1); Flavobacterium spp. (14, 46, 2); Fusobacterium spp. (13, 1, 1); Haemophilus spp. (25, 86, 5); Hafnia spp. (25, 7, 5); Listeria spp. (8, 2, 1); Micrococcus spp. (40, 88, 6); Moraxella spp. (12, 18, 2); Morganella spp. (37, 19, 7); Neisseria spp. (23, 2, 1); Nocardia spp. (17, 127, 1); Oligella spp. (18, 12, 2); Peptostreptococcus spp. (25, 10, 2); Providencia spp. (19, 6, 4); Stenotrophomonas spp. (28, 346, 3) |
| Zoonotic infection | Yersinia spp. (3, 8, 1) |
Frequency of Reported Antibiotic Resistance or Susceptibility in Iranian Studies Based on the Type of Bacteria
3.2. Characteristics of Included Studies
This review assessed the quality of reporting in terms of the main characteristics of each study. The study information was insufficient in 306 studies. The missing information was identified for the year and the city of studies in 245 and 58 reports, respectively. Only 193 studies (11.2%) reported the age characteristics of the study population.
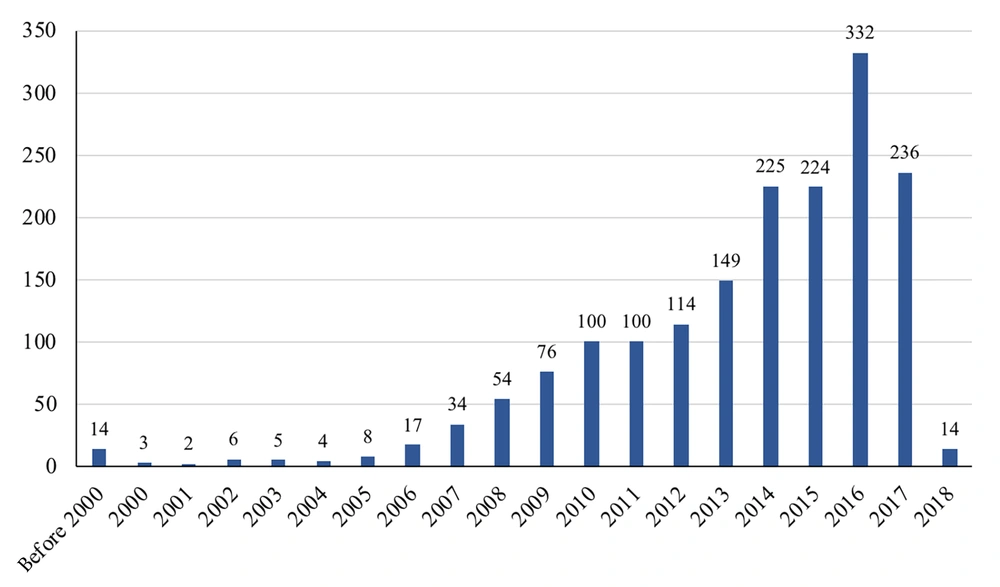
The years of publication for the included studies are shown in Figure 2. Accordingly, 59.2% of the studies were published within 2014 and 2017, with the highest number of publications in 2016. The interval between the end of studies and publications in scientific journals was diverse, ranging from less than 1 year to 19 years. Among 1,473 studies that reported the study year, 1,242 studies (84.3%) published their results in 3 years or less, 196 studies between 4 and 5 years, 25 studies between 6 and 7 years, 2 studies after about 8 years, 6 studies between 10 to 12 years, and 1 study after 19 years. Most of the studies were conducted after 2010 (72.8%). The pattern of publication year for each common or specific bacterium is shown in Table 3.
| Variables | Before 2000 | 2000 - 2004 | 2005 - 2009 | 2010 | 2011 | 2012 | 2013 | 2014 | 2015 | 2016 | 2017 | 2018 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acinetobacter baumannii | 9 | 9 | 10 | 13 | 20 | 28 | 28 | 35 | 36 | 2 | ||
| Alpha-hemolytic Streptococcus | 1 | 2 | 2 | 1 | ||||||||
| Bacillus cereus | 1 | |||||||||||
| Bacteroides fragilis | 1 | 1 | 1 | |||||||||
| Beta-hemolytic Streptococcus | 2 | 1 | 1 | 1 | 1 | 1 | ||||||
| Bordetella pertussis | 1 | 1 | ||||||||||
| Brucella spp. | 2 | 2 | 1 | 4 | 1 | |||||||
| Burkholderia spp. | 1 | |||||||||||
| Campylobacter spp. | 1 | 4 | 3 | 1 | 2 | 1 | ||||||
| Clostridium difficile | 1 | 1 | 1 | 1 | ||||||||
| Clostridium perfringens | 2 | |||||||||||
| Coagulase-negative staphylococci | 1 | 1 | 28 | 14 | 8 | 13 | 19 | 25 | 30 | 31 | 28 | 2 |
| Enterobacter spp. | 2 | 1 | 29 | 9 | 7 | 16 | 19 | 20 | 15 | 33 | 17 | |
| Enterococcus faecalis | 1 | 16 | 3 | 1 | 3 | 4 | 8 | 4 | 21 | 9 | 2 | |
| Enterococcus faecium | 18 | 2 | 1 | 2 | 3 | 6 | 4 | 17 | 7 | 3 | ||
| Enterococcus spp. | 22 | 8 | 5 | 13 | 9 | 11 | 18 | 26 | 12 | 1 | ||
| Escherichia coli | 4 | 2 | 52 | 30 | 28 | 34 | 48 | 67 | 55 | 90 | 58 | 2 |
| Haemophilus influenza | 1 | 4 | 2 | 3 | 1 | 2 | 1 | 2 | ||||
| Helicobacter pylori | 2 | 7 | 5 | 8 | 5 | 8 | 9 | 4 | 7 | 3 | ||
| Klebsiella pneumoniae | 1 | 18 | 22 | 10 | 15 | 17 | 35 | 23 | 43 | 27 | 2 | |
| Legionella pneumophila | 1 | 1 | ||||||||||
| Listeria monocytogenes | 1 | 1 | 1 | 1 | 1 | |||||||
| Moraxella catarrhalis | 1 | 1 | ||||||||||
| Mycobacterium tuberculosis | 1 | 4 | 16 | 3 | 3 | 2 | 4 | 12 | 10 | 2 | ||
| Mycoplasma spp. | 1 | |||||||||||
| Neisseria gonorrhoeae | 1 | 1 | 1 | 1 | 1 | |||||||
| Neisseria meningitidis | 2 | 1 | 1 | 1 | 1 | |||||||
| Nonhemolytic streptococci | 1 | 1 | 2 | 1 | 1 | 3 | ||||||
| Nontuberculous mycobacteria | 1 | 1 | 1 | 2 | 2 | |||||||
| Porphyromonas gingivalis | 1 | |||||||||||
| Propionibacterium acnes | 1 | 1 | ||||||||||
| Pseudomonas aeruginosa | 2 | 4 | 39 | 22 | 16 | 25 | 29 | 48 | 38 | 58 | 40 | |
| Salmonella enterica | 1 | 3 | 4 | 1 | 1 | 1 | 5 | 1 | ||||
| Salmonella spp. | 3 | 6 | 4 | 2 | 3 | 1 | 6 | 4 | 6 | 7 | ||
| Salmonella typhi | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 2 | 1 | |||
| Salmonella typhimurium | 1 | 1 | 4 | 2 | ||||||||
| Shigella dysenteriae | 2 | 1 | 9 | 2 | 1 | 2 | 1 | 3 | 2 | 3 | ||
| Shigella flexneri | 2 | 1 | 10 | 3 | 2 | 3 | 3 | 3 | 1 | 5 | 5 | |
| Shigella sonnei | 2 | 1 | 10 | 5 | 1 | 2 | 3 | 4 | 1 | 6 | 6 | 1 |
| Shigella spp. | 4 | 1 | 13 | 2 | 1 | 3 | 5 | 2 | 1 | 6 | 5 | |
| Staphylococcus aureus | 2 | 4 | 61 | 22 | 17 | 26 | 41 | 48 | 57 | 72 | 48 | 5 |
| Stenotrophomonas maltophilia | 1 | 1 | 1 | 1 | 1 | 4 | 3 | |||||
| Streptococcus agalactiae | 4 | 1 | 1 | 2 | 1 | 3 | 3 | 6 | 2 | 2 | ||
| Streptococcus pneumoniae | 2 | 2 | 11 | 6 | 3 | 7 | 5 | 11 | 5 | 6 | 4 | 1 |
| Streptococcus pyogenes | 1 | 1 | 3 | 1 | 2 | 3 | ||||||
| Streptococcus viridans | 5 | 2 | 1 | 1 | 2 | 1 | 1 | |||||
| Vibrio cholerae | 2 | 3 | 1 | 3 | 1 | 2 | 2 | 5 | 1 | |||
| Yersinia spp. | 1 |
Number of Studies for Clinically Important Bacterium According to the Year of Publication
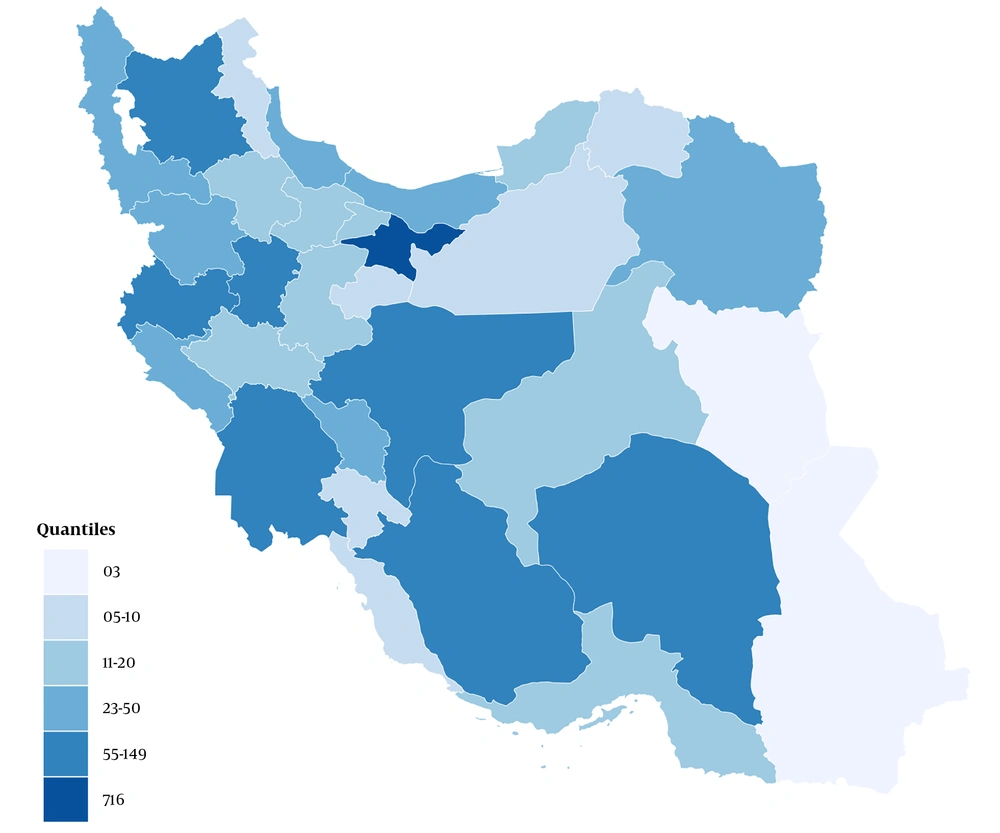
Tehran province, with 716 studies, had the highest number of study locations. Isfahan, Fars, and East Azarbaijan provinces, with 149, 108, and 104 studies, had the second to fourth rank in terms of the number of study sites, respectively. The distribution of the provinces of studies is shown in Figure 3.
The included studies have been published in 376 scientific journals, 32.4% of which were local. Jundishapur Journal of Microbiology had the highest number of publications, with 131 papers; however, 163 journals published only one study. Moreover, 110 retrieved studies were published as conference proceedings only. The list of journals with 20 or more publications (71.4% in the local journals) is provided in Table 4.
| Name of Journal | Journal Impact Factor (2021) a | No of Publications |
|---|---|---|
| Jundishapur Journal of Microbiology | 0.747 | 131 |
| Iranian Journal of Public Health | 1.291 | 60 |
| Iranian Journal of Microbiology | 1.221 | 53 |
| Archives of Pediatric Infectious Diseases | - | 40 |
| Journal of Isfahan Medical School | - | 35 |
| African Journal of Microbiology Research b | - | 33 |
| Journal of Mazandaran University of Medical Sciences | - | 31 |
| Microbial Drug Resistance b | 3.431 | 24 |
| Archives of Clinical Infectious Diseases | - | 23 |
| Iranian Red Crescent Medical Journal | - | 23 |
| Microbial Pathogenesis b | 3.738 | 23 |
| Iranian Journal of Clinical Infectious Diseases | - | 21 |
| Iranian Journal of Medical Microbiology | - | 21 |
| Journal of Pure and Applied Microbiology b | - | 21 |
Journals with the Highest Number of Publications
4. Discussion
The increasing rate of scientific publications related to AMR makes the synthesis of scientific knowledge more noteworthy than ever. In light of this issue, this scoping review aimed to map the existing studies on AMR clinical research in Iran and identify quality gaps in reporting.
The present analysis showed that AMR was not a focus of interest among Iranian researchers by 2005. However, since 2006, the number of publications gradually surged; accordingly, in 2016, an unprecedented volume of research on this topic was published. It is worth mentioning that the data for the year 2018 are not completed, as the study period was closed in February 2018. The increasing pattern of Iranian publications on AMR is following the global trend of studies on this topic, which could be due to the growing burden of AMR in recent years (2, 11).
According to the global burden of disease (GBD) studies on AMR, 6 pathogens were responsible for 929,000 deaths attributable to AMR in 2019: Escherichia coli, followed by Staphylococcus aureus, K. pneumoniae, S. pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa (12). As shown in Table 3, researchers’ tendency to study gram-negative bacteria, which cause a high burden of diseases, has increased in recent years. In line with the GBD studies on AMR, Iranian researchers are more likely to evaluate the resistance of E. coli and K. pneumonia to the third generation of cephalosporins and Mycobacterium tuberculosis to isoniazid. However, unlike the assessments of GBD that highlighted the burden of resistance of Shigella spp. to fluoroquinolones, S. pneumoniae to penicillin, S. aureus to methicillin, Salmonella typhi and paratyphi to fluoroquinolones and chloramphenicol, and Neisseria gonorrhoeae to the third generation of cephalosporins, the Iranian publications were more frequently focused on evaluating the resistance of Shigella spp. to cotrimoxazole, S. pneumoniae to gentamicin, S. aureus to ofloxacin, S. typhi, and paratyphi to ampicillin and cotrimoxazole, and N. gonorrhoeae to penicillin (2).
This study also analyzed the quality of reporting in terms of the key characteristics of the studies. The first finding was that the duration between the completion of a study and publication ranged from less than 1 year to about 19 years, and for most of the publications, it was around 3 years. This delay can be caused by several factors depending on authors and editors. The factors related to authors are listed as delays in data analysis, manuscript preparation and submission, and responding to editorial comments. Journals-related factors are also identified as journal rejections and the editorial process (13). The publication lag time might affect treatment choices and the prioritization of future research (14).
The low quality of reporting was detected in 27.2% of the studies concerning the participant information, the age of the study population, the year of study, the city of study, and the number of impaired participants. The problems in reproducibility and transferability of the publications cause the shift in researchers’ preferences to publish their manuscripts in low-impact factor (IF) journals (15, 16). It was determined that there is a gap in the quality of reporting of AMR intervention research, which represents a challenge for interpreting and replicating the research findings (17). The absence of a comprehensive surveillance process for AMR is reported in low- and middle-income countries (18), which might serve as an alternative to the large number of small studies that are currently being conducted.
The present review mapped the vast majority of studies; however, there are a few limitations, such as the fact that the national IDML database has not been updated and has not been functioning properly since 2021, which means it was impossible to update the current search accordingly. Furthermore, it is pointed out that the number of studies performed on each antibiotic, bacterium, or location should not be interpreted as a proxy for AMR.
4.1. Conclusions
This scoping review summarizes the literature on AMR research in Iran to provide an account of the current research and guide future research priorities. This review showed that despite the extensive research on AMR in Iran, the quality of reporting needs further improvement. Given the significance of addressing the threat of AMR, it is essential for health research governance to prioritize conducting timely, effective, and well-reported surveillance of AMR. In addition to promoting the quality of evidence, such surveillance can prevent the waste of research resources.