1. Background
Patients requiring intensive care unit (ICU) admission are prone to nosocomial infections (NIs), five to seven-fold more compared to general hospital wards (1). In spite of significant changes in the spectrum of microorganisms causing ICU-associated NIs, Pseudomonas aeruginosa has held a nearly unchanged position in the rank order of pathogens causing ICU-related NIs during the last four decades, especially among patients, who have undergone mechanical ventilation (2-4). Furthermore, ventilator-associated pneumonia (VAP) caused by P. aeruginosa remains a severe and dreaded complication. Risk of mortality and morbidity in VAP is increased due to wrong or delayed initial antibiotic therapy, especially when VAP is caused by multidrug-resistant pathogens (5). Eighty-six percent of nosocomial cases of pneumonia are associated with VAP. The mortality due to VAP has been reported to range between 0% and 50% (6). Compared with community-acquired strains, nosocomial P. aeruginosa isolates tend to be more resistant (2). Nowadays, antimicrobial resistance is a major problem for the treatment and management of NIs, especially for patients admitted to ICUs (7). Patients with drug-resistant organisms are at risk of negative outcome. Pseudomonas aeruginosa is naturally resistant to many antimicrobial agents and it can also acquire resistance against available antibiotics through multiple mechanisms (8). Furthermore, ESBL-producing bacteria cause serious infections and have high mortality rates. ESBL-bacteria are capable of efficiently hydrolyzing many beta-lactam antibiotics, including cephalosporins and monobactams (9, 10). The most commonly encountered ESBLs are derived from the sulf-hydryl variable (SHV), cefotaxime-beta lactamases (CTX), vietnam extended-spectrum β-lactamase (VEB), and Guyana extended-spectrum ß-lactamases (GES) genes (11-14). Integrons are one of the mobile genetic elements, which are able to carry genes for resistance to different antibiotics. Integrons are divided to four classes based on the type of integrase genes. Resistance genes, which are located in the gene cassettes, can be separated and entered to other integrons. This is an important phenomenon in the creation and distribution of new resistance cassettes. Importance of association of multidrug resistance and presence of integron plays an important role in the development of multiple resistance (15-17). A shift in the distribution of ESBL-producing strains has recently occurred with reports from North America, Europe, South America, Africa, and Asia, with a dramatic increase of CTX, TEM, and SHV variants (18-21). Also, the incidence of ESBL-producing strains has increased in different geographic regions of Iran, such as north of Iran (9, 10, 22-32).
2. Objectives
Designing a control program will require insight of a range of disciplines, including epidemiology, molecular biology, and evolutionary biology of resistance genes. Regarding the clinical importance of class 1 and 2 integrons in antibiotic resistance and considering that there are no reports available on prevalence of integrons as mobile genetic elements carrying antibiotic resistance gene cassette in ESBL P. aeruginosa isolated from ventilator-associated NIs in ICU wards of northern Iran, this study attempted to screen for ESBL genes, and confirm their resistant characteristics.
3. Methods
3.1. Study Population and Specimen Types
This study was conducted at 18 hospitals in Mazandaran province, north of Iran, with 1200 ward beds and 100 intensive care unit beds during years 2014 and 2015. This study was approved by the ethics committee of Mazandaran University of Medical Sciences (Code No: 879 Date: July 9, 2014). For all patients, samples were taken within 48 hours of admission from mechanical ventilation. Non-duplicate isolates of P. aeruginosa were collected from various specimens of all patients with microbiologically confirmed ventilator-associated NIs. All patients aged 18 years and older with ventilation for at least 48 hours were assessed daily for evidence of ventilator-associated NIs. Patients, who were chronically mechanically ventilated were excluded. Only microbiologically confirmed episodes of VAP were considered for analysis.
3.2. Microbiological Methods
All samples were routinely cultured on MacConkey and blood agar plates. Blood samples were cultured in blood culture bottles. Isolates were identified at the species level using standard biochemical tests and microbiological methods (33).
3.3. Antibiotic Susceptibility
Susceptibility of the clinical isolates to routinely used antibiotics was determined by the standard broth dilution (microdilution broth) technique. The minimal inhibitory concentrations (MIC) was determined according to the recommendations of clinical and laboratory standards institute 2010 (CLSI). The antibiotics that were used were amikacin, ciprofloxacin, imipenem, gentamicin, ceftazidime, tobramycin, piperacillin-tazobactam, cefepime, colistin, and co-trimoxazole. The antibiotics were purchased from Sigma chemical company.
3.4. Detection of ESBL-Producing Pseudomonas aeruginosa
To screen for ESBL-producing strains with MIC of > 8 μg/mL to cephalosporins, at least one of them including cefotaxime, cefepime, ceftazidime, and ceftriaxone were tested using the double-disk synergy test. The presence of ESBL was assayed, using the following antibiotic disks: cefotaxime (30 μg), cefotaxime/clavulanic acid (30/10 μg), ceftazidime (30 μg), and ceftazidime/clavulanic acid (30/10 μg) (MAST, UK). Escherichia coli ATCC 25922 strains served as positive controls (9, 25, 34).
3.5. DNA Isolation and Genotyping
Bacterial DNA was extracted using a commercial gene extraction kit (Takapou Zist, Iran), according to the company’s recommendation. Then, ESBL-positive strains were screened for CTX, VEB, GES, SHV, and integron class 1 and class 2 genes performed by PCR amplification. The primer sequences and PCR annealing temperature are shown in Table 1. In all experiments, the following reference strains were used as positive controls: K. pneumoniae 7881 (CTXM), K. pneumoniae 7881 strain (containing SHV), P. aeruginosa ATCC 27853 (VEB-1), and K. pneumoniae (containing GES), which were kindly provided by Professor P. Nordmann CHU Bicetre, France. Escherichia coli 96K062 was used as a positive control for class 1 and 2 integrons. A non-ESBL-producing strain (E. coli ATCC 25922) was used as a negative control.
| Target Genes | Primer Used (5’ - 3’) | Ref | Thermal Cycling Condition | PCR Product Size |
|---|---|---|---|---|
| CTX | TTTGCGATGTGCAGTACCAGTAA | (35) | 94°C 5 min → 40 × [94°C 45 sec, 53.1°C 45 sec, 72°C 1 min] → 72°C 7 min | 593 bp |
| CGATATCGTTGGTGGCATA | ||||
| VEB | CGACTTCCATTTCCCGATGC | (36) | 93°C 3 min → 40 × [93°C 1 min, 54.9°C 1 min, 72°C 1 min] → 72°C 7 min | 585 bp |
| GGACTCTGCAACAAATACGC | ||||
| GES | ATGCGCTTCATTCACGCAC | (37) | 1 cycle of 5 min at 95°C; 30 cycles of 1 min at 95°C, 45 sec at 55°C, 30 sec at 72°C; 1 cycle of 8 min at 72°C | 846 bp |
| CTATTTGTCCGTGCTCAGG | ||||
| SHV | AAGATCCACTATCGCCAGCAG | (38) | 1 cycle of 3 min at 94°C; 35 cycles of 30 sec at 94°C, 1 min at 60°C, 1 min at 72°C; 1 cycle of 7 min at 72°C | 231 bp |
| ATTCAGTTCCGTTTCCCAGCGG | ||||
| INT1 | CAGTGGACATAAGCCTGTTC | (39) | 1 cycle of 2 min at 94°C; 35 cycles of 30 sec at 94°C, 30 sec at 55°C, 30 sec at 72°C; 1 cycle of 3 min at 72°C | 160 bp |
| CCCGAGGCATAGACTGTA | ||||
| INT2 | TTGCGAGTATCCATAACCTG | (40) | 1 cycle of 5 min at 95°C; 30 cycles of 45 sec at 94°C, 40 sec at 58°C, 1 min at 72°C; 1 cycle of 7 min at 72°C | 288 bp |
| TTACCTGCACTGGATTAAGC |
3.6. Statistical Analysis
The SPSS software version 16 and descriptive tests, such as frequency analysis and median value, were used for statistical analysis.
4. Results
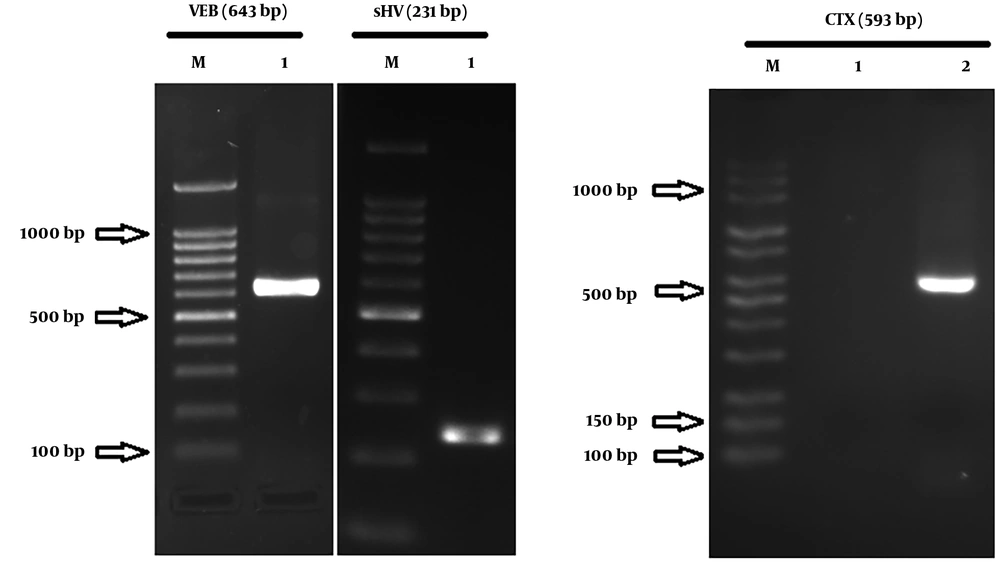
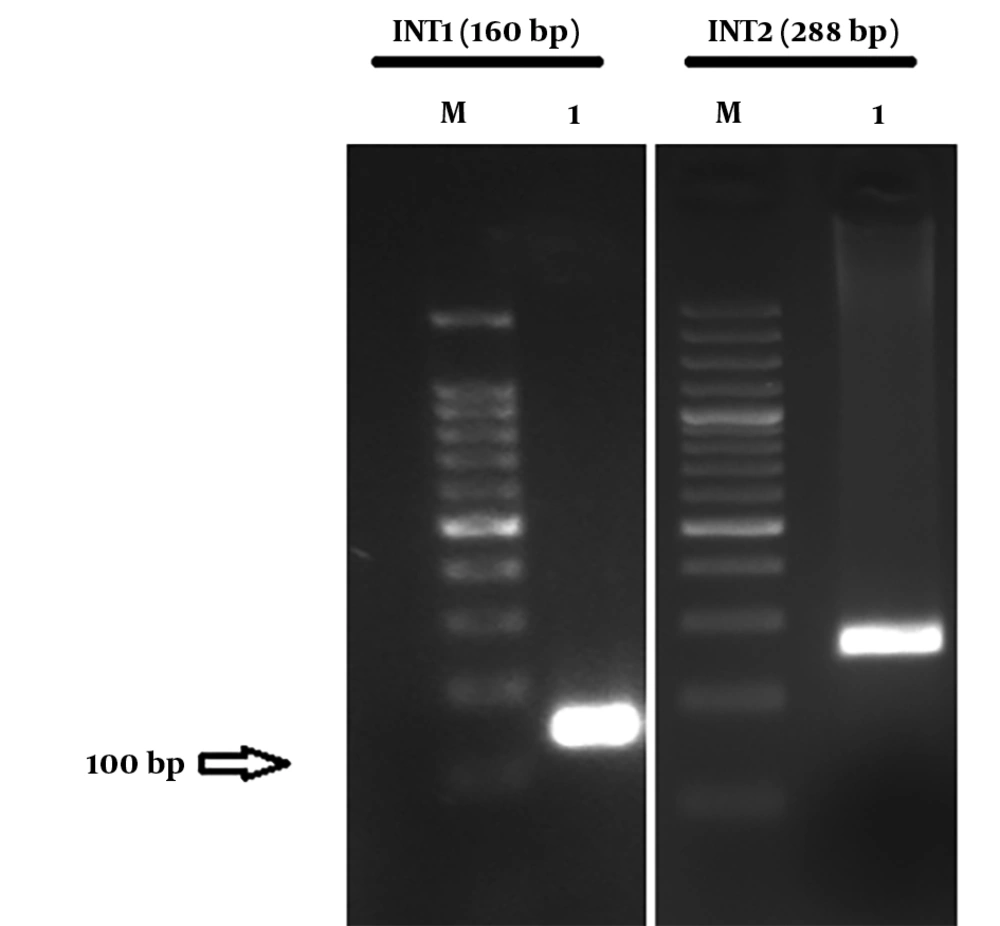
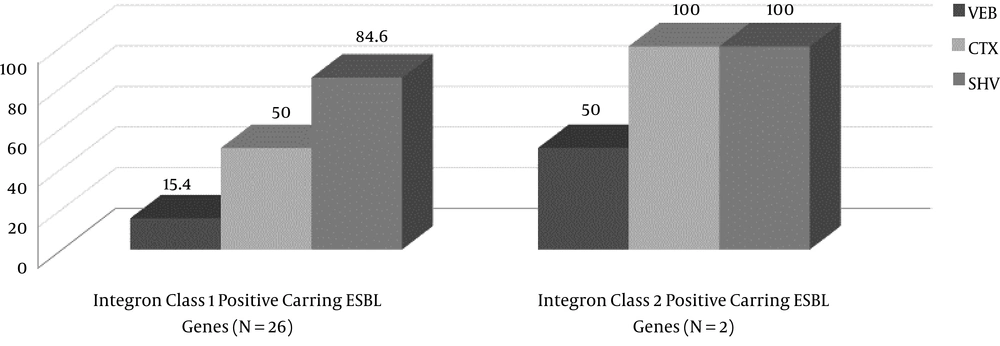
From a total of 205 hospitalized patients with NIs at ICUs of eighteen hospitals during years 2014 and 2015, thirty patients had ventilator-associated nosocomial infection caused by ESBL-producing P. aeruginosa. Seventeen patients (56.7%) were male and 13 (43.3%) were female. The average age was 54.47 ± 30 years old. The average duration of hospitalization at ICU wards was 29 ± 13.5 days. The prevalence of ventilator-associated infection for VAP was (25 patients) 83.33%; also, 16.6% (five patients) had sepsis due to VAP. Distribution of CTX, VEB, and SHV genes in ESBL-producing P. aeruginosa was 13 (43.33%), four (13.33%), and 26 (86.66%), respectively. The ESBL-producing P. aeruginosa carrying integron class 1 and class 2 were 26 (86.66%) and two (6.66%), respectively. Figure 1 and 2 show the agarose gel of the strains containing ESBL and Integrons gene, respectively. Fourteen strains contained the SHV gene, thirteen strains contained two ESBL genes (10 strains had CTX and SHV, two strains had VEB and SHV, and one strain had VEB and CTX), two strains contained three ESBL genes (VEB, CTX, and SHV), and one strain had only CTX gene. The presence or absence of ESBL genes, susceptibility, and resistance to different antimicrobial agents for ESBL-related genes and Integron class 1 and 2 are shown in Table 2 and 3, respectively. Two isolates (6.66%) had both classes of integrons, simultaneously. The incidence of ESBL genes in intergron positive isolates is illustrated in Figure 3. None of isolates had the GES gene.
| Gene | ||||||||
|---|---|---|---|---|---|---|---|---|
| SHV | VEB | CTX | GES | |||||
| Negative, N = 4 | Positive, N = 26 | Negative, N = 26 | Positive, N = 4 | Negative, N = 17 | Positive, N = 13 | Negative, N = 30 | Positive, N = 0 | |
| Amikacin | 0 | |||||||
| R | 100 | 69.23 | 73.07 | 75 | 76.47 | 69.23 | 73.33 | |
| I | 0 | 15.38 | 15.38 | 0 | 11.76 | 15.38 | 13.33 | |
| S | 0 | 15.38 | 11.53 | 25 | 11.76 | 15.38 | 13.33 | |
| Ciprofloxacin | 0 | |||||||
| R | 50 | 46.15 | 46.15 | 50 | 64.70 | 23.07 | 46.66 | |
| I | 25 | 23.07 | 23.07 | 25 | 11.76 | 38.46 | 23.33 | |
| S | 25 | 30.76 | 30.76 | 25 | 23.52 | 38.46 | 30 | |
| Imipenem | 0 | |||||||
| R | 25 | 26.92 | 23.06 | 50 | 35.29 | 15.38 | 26.66 | |
| I | 50 | 15.38 | 19.23 | 25 | 23.52 | 15.38 | 20 | |
| S | 25 | 57.69 | 57.69 | 25 | 41.17 | 46.15 | 53.33 | |
| Gentamicin | 0 | |||||||
| R | 75 | 88.46 | 84.61 | 100 | 82.35 | 92.30 | 86.66 | |
| I | 0 | 0 | 0 | 0 | 0 | - | - | |
| S | 25 | 11.53 | 15.38 | 0 | 17.64 | 7.69 | 13.33 | |
| Ceftazidime | 0 | |||||||
| R | 100 | 61.53 | 65.38 | 75 | 70.58 | 61.53 | 66.66 | |
| I | 0 | 26.92 | 23.07 | 25 | 23.52 | 23.07 | 23.33 | |
| S | 0 | 11.53 | 11.53 | 0 | 5.88 | 15.38 | 10 | |
| Tobramycin | 0 | |||||||
| R | 50 | 46.15 | 38.46 | 100 | 41.17 | 53.84 | 46.66 | |
| I | 25 | 23.07 | 26.92 | 0 | 29.41 | 15.38 | 23.33 | |
| S | 25 | 30.76 | 34.61 | 0 | 29.41 | 30.76 | 30 | |
| Piperacillin-Tazobactam | 0 | |||||||
| R | 75 | 84.61 | 80.76 | 100 | 82.23 | 76.92 | 83.33 | |
| I | 25 | 15.38 | 19.23 | 0 | 11.76 | 23.07 | 16.66 | |
| S | 0 | - | - | 0 | - | - | - | |
| Cefepime | 0 | |||||||
| R | 100 | 76.92 | 92.30 | 75 | 100 | 53.84 | 76.66 | |
| I | 0 | 19.23 | 15.38 | 25 | 0 | 38.46 | 16.66 | |
| S | 0 | 3.84 | 3.84 | 0 | 0 | 7.69 | 3.33 | |
| Colistin | 0 | |||||||
| R | 50 | 57.69 | 53.84 | 75 | 70.58 | 38.46 | 56.66 | |
| I | 50 | 26.92 | 30.76 | 25 | 23.52 | 38.46 | 30 | |
| S | - | 15.38 | 26.92 | 0 | 5.88 | 23.07 | 13.33 | |
| Co-trimoxazole | 0 | |||||||
| R | 100 | 88.46 | 84.61 | 100 | 94.11 | 76.92 | 86.66 | |
| I | - | 3.84 | 3.84 | 0 | 0 | 7.69 | 3.33 | |
| S | - | 11.53 | 11.53 | 0 | 5.88 | 15.38 | 10 | |
Abbreviations: R, resistant; I, intermediate; S, sensitive.
a Numbers in table are in percentage.
| Gene | ||||||
|---|---|---|---|---|---|---|
| Antibiotics | Integron Class 1 Positive, N = 26 | Integron Class 2 Positive, N = 2 | ||||
| R | I | S | R | I | S | |
| Amikacin | 76.9 | 11.5 | 11.5 | 50 | 50 | 0 |
| Ciprofloxacin | 42.3 | 26.9 | 30.8 | 100 | 0 | 0 |
| Imipenem | 23.1 | 23.1 | 53.8 | 50 | 0 | 50 |
| Gentamicin | 88.5 | 0 | 11.5 | 100 | 0 | 0 |
| Ceftazidime | 69.2 | 19.2 | 11.5 | 50 | 50 | 0 |
| Tobramycin | 50 | 15.4 | 34.6 | 50 | 50 | 0 |
| Piperacillin-Tazobactam | 80.8 | 19.2 | 0 | 100 | 0 | 0 |
| Cefepime | 76.9 | 19.2 | 3.8 | 50 | 50 | 0 |
| Colistin | 57.7 | 30.8 | 11.5 | 50 | 50 | 0 |
| Co-trimoxazole | 88.5 | 3.8 | 7.7 | 100 | 0 | 0 |
Abbreviations: R, resistant; I, intermediate; S, sensitive.
a Numbers in table are in percentage.
5. Discussion
Culprits of late VAP are typically MDR bacteria, such as ESBL bacteria (41). This research found that ESBL P. aeruginosa was responsible for 36.5% of VAP at ICUs of 18 hospitals in the north of Iran, during years 2014 to 2015. Pseudomonas aeruginosa with a frequency of 24.4% is the most important pathogen causing VAP (42-44). Similar to the current findings, Gupta et al. reported that Pseudomonas spp. with incidence of 35 (28%) was the most prevalent pathogen isolated from patients admitted in ICU. The similarity of the results of the current study and Gupta et al.’s findings may be explained by the homology of patients of the examined wards of the hospitals. On the other hand, the most important risk factor significantly associated with infections caused by Pseudomonas spp. at ICU in Gupta et al.’s study was mechanical ventilation, similar to the current study (45); although VAP spreads to the blood in 10% of cases (6). This study found 16.6% sepsis due to VAP caused by ESBL P. aeruginosa. Pelekanou et al. investigated the differences in innate and adaptive immune responses in 36 patients with sepsis and VAP and 32 patients with sepsis due to other infections. They found more pronounced immunoparalysis in patients with VAP than in those with other bacterial infections. This was supported by the decreased number of CD3+/CD4+ cells, the increase in monocyte apoptosis, and the lower release of pro-inflammatory cytokines, namely tumor necrosis factor-alpha, and interleukin-6, from monocytes after stimulation with lipopolysaccharide in the group of patients with VAP (46-48).
The current study revealed that on average 26.6% to 86.66% of resistance to routine antibiotics used for infection, are caused by P. aeruginosa. Imipenem with 53.3% sensitivity was the most effective antibiotic. In some studies conducted in developed countries, the rate of antibiotic resistance to beta-lactam antibiotics and fluoroquinolones of P. aeruginosa was lower than the current results (49-51). In this regard, it is necessary to avoid prescribing unnecessary antibiotics, intravenous administration of short-term antibiotics to prevent infection in high-risk patients, and conservative use of medical equipment such as ventilator in developed countries (22).
In P. aeruginosa, various classes of ESBLs (A, B and D) have been found. Five types of class A ESBLs (PER, VEB, GES and IBC, TEM and SHV) were recently reported (38). The prevalence of different ESBL genotypes varies in different countries. The VEB, GES , SHV, and CTX genotypes are more prevalent in Asian countries, thus, the presence of these genes was evaluated in the current study (52).
Although more than 300 different ESBL variants have been described, TEM and SHV variants were the most common ESBLs during the past decade; strains expressing CTX have begun to emerge in many regions (53, 54).
This study found that the most prevalent gene for ESBL production was SHV, which was detected in 26 (86.6%) isolates. It has been proven that prevalence of ESBL genes varies in different geographic regions, for example in line with the current study, Imani Foolad et al. (55), reported that the most common ESBL gene detected among P. aeruginosa isolates in Tehran hospitals was SHV (37.5%). On the other hand, Bokaeian et al. (56), in Zahedan found that TEM was the most prevalent ESBL gene and only 6.6% of P. aeruginosa isolates contained the SHV gene.
The incidence of CTX gene among carbapenem resistant isolates in the current study was estimated about 16% and on the other hand, SHV and VEB had rates of 26.9% and 50% among imipenem resistance P. aeruginosa in the current study, which indicates the critical therapeutic problem of NIs caused by P. aeruginosa.
Furthermore, VEB and GES genes as plasmid-mediated ESBLs are uncommon and have been found mainly in P. aeruginosa. The VEB-type beta-lactamases are a rare type of enzyme responsible for conferring ESBL; they are usually integron borne and horizontally transferred (53). The VEB enzymes are well inhibited by clavulanate, and lead to significant reductions in ceftazidime. Thirteen percent of isolates in the current study had the VEB gene and the MIC showed that antibiotic resistance rate among VEB-containing isolates was 50% to 100%, and cefepime and ceftazidime were the most resisted antibiotics and imipenem was the most sensitive antibiotic. Similar to the current results, Davoudian et al. (54), reported that among 10 ESBL-positive ICU P. aeruginosa, one (10%) isolate contained the VEB gene yet 50% of the ESBL isolates in their study were resistant to third generation cephalosporins, indicating that the presence of other ESBL enzymes, metallo-beta-lactamases or mechanisms including efflux pumps for cephalosporin resistance may be cooperated in this phenomenon.
In the current study, 46% of fluoroquinolone-resistant isolates had the SHV gene. Horizontal transfer of genes encoding acquired beta-lactamases, such SHV, seems to play a primary role in the dissemination of these resistance traits among fluoroquinolone-resistant clinical strains of P. aeruginosa. The incidence of CTX and SHV, simultaneously, was the most prevalent after SHV in the current study.
Similar to the current findings, the GES gene was not seen in P. aeruginosa strains in several studies of Iran. The GES gene plays an important role in resistance to carbapenem antibiotics yet the absence of this gene in imipenem resistant isolates in these studies could be explained by other mechanisms of acquiring carbapenem-resistant phenotype, such as other ESBL enzymes (57-59).
This study found that 86.6% and 6.6% of isolates were Integron class 1 and class 2 positive. About 15.4% to 85% of integron class 1 positive strains contained ESBL genes and the incidence of ESBL genes was associated with the presence of integrons. In fact, integrons are expression vectors for antibiotic resistance genes that are included as gene cassettes (60). Mobile elements, such as integrons, may facilitate the spread of ESBL genes among bacteria. The rate of class 1 integron in several studies was around 40% for P. aeruginosa (61-64). It seems that the prevalence of integrons in the current study was higher than other studies, which can be due to differences in the prevalence of class 1 integron gene in different geographic areas, the number, and type of samples. As this study only evaluated the ESBL P. aeruginosa, the higher rate of integron positivity in comparison with other studies is not surprising. The correct identification of the genes involved in ESBL-mediated resistance is necessary for the surveillance and epidemiological studies of their transmission (65). Molecular typing of ESBL-producing P. aeruginosa, such as pulsed-field gel electrophoresis (PFGE) or multilocus sequence typing (MLST) is useful for surveillance purposes and is recommended for future study. The result of this study was alarming for clinicians to consider the risk of healthcare-associated pneumonia, such as pneumonia caused by ESBL-producing bacteria in patients admitted to ICUs.