1. Background
After the emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in late December 2019, coronavirus disease 2019 (COVID-19) has turned into a nightmare, with over six million recorded deaths since the beginning of the pandemic (1). Scientists worldwide have been uncovering new aspects of SARS-CoV-2 immunopathogenesis to identify appropriate therapeutic and prophylactic approaches (2). The immune responses to SARS-CoV-2 are primarily initiated by infected alveolar macrophages and lung epithelial cells within the innate branch of immunity, which subsequently activates T and B cells in the adaptive immune system (3). Indeed, the nature and intensity of the immune responses to SARS-CoV-2 determine the disease outcome (3).
SARS-CoV-2 hampers the production of antiviral interferon (IFN) by infected ACE2-expressing cells in the lungs. However, adaptive immune cells recruited to the lungs amplify inflammatory responses, worsening the clinical outcomes of COVID-19 patients (4). Nonetheless, the induction of a humoral response could partially control virus spread through antibodies that primarily target the spike glycoprotein and the nucleocapsid protein of SARS-CoV-2. Mainly, neutralizing antibodies (NAs) to the SARS-CoV-2 receptor binding domain (RBD) play a crucial role in viral neutralization (2). In this context, most COVID-19 patients undergo seroconversion between 7 - 14 days after the onset of symptoms, and antibody levels persist for several weeks, contributing to viral eradication (5, 6).
Padoan et al. demonstrated that IgM specific to the SARS-CoV-2 spike protein peaked at 10 - 12 days after symptom onset and later declined after 18 days (7). In a cohort study, the peak of IgM occurred 20 - 22 days after symptom onset, while IgG seroconversion was observed in all COVID-19 patients within 17 - 19 days after symptom onset (6). Liu et al. reported that anti-SARS-CoV-2 spike-specific IgM became detectable from day 4 onward, peaking at approximately day 20, and then gradually decreasing. Furthermore, anti-SARS-CoV-2 spike-specific IgGs were detectable from day 7 onwards, reaching their peak at approximately day 25 (8). Another study indicated that SARS-CoV-2 spike IgG titers increased during the first 3 weeks after the onset of COVID-19 symptoms and began to decline after 8 weeks (9). Overall, there is no consistency in the timing and profile of anti-SARS-CoV-2 antibodies across published papers.
Additionally, the humoral response to SARS-CoV-2 infection may differ from that elicited by vaccination (10-13). In this regard, the pattern of IgG to SARS-CoV-2 spike and RBD, as well as NAs in COVID-19-infected individuals and vaccine recipients, could provide valuable insights into evaluating immune responses to the virus. Therefore, we aim to assess the pattern of IgG antibodies to spike and RBD, along with neutralizing antibodies, in vaccine recipients and COVID-19-infected patients to gain a better understanding of the serological profile of the humoral response in COVID-19.
2. Objectives
This study aims to investigate the seroconversion patterns of specific antibodies to various antigens of SARS-CoV-2 in individuals hospitalized due to COVID-19 and those who have received vaccinations. The main emphasis will be on comparing the humoral responses between individuals who have been infected with the virus and those who have been vaccinated.
3. Methods
3.1. Study Design and Participants
In this retrospective cohort study, a total of 134 patients, admitted to Masih Daneshvari Hospital between April 20, 2020, and August 20, 2020, with COVID-19, were enrolled. Upon admission, nasopharyngeal and oropharyngeal swab samples and blood were collected from each patient. The gold standard test for diagnosing SARS-CoV-2 and the inclusion criteria was the real-time RT-PCR test. Among all the enrolled patients, 102 were confirmed cases of COVID-19 based on positive molecular testing and clinical findings (Appendices 1 and 2 in the Supplementary File). Serial blood and swab samples were collected during hospitalization and after discharge on days 0, 3, 7, 14, 21, and 36 post-admission. Additionally, 141 serum samples were collected from vaccinees between June 20, 2021, and July 20, 2021, after receiving either the first or second dose of different vaccines (Appendix 3 in the Supplementary File).
3.2. RNA Extraction and RT-PCR Assay
Total RNAs were extracted from patients' swabs using a column-based kit as per the manufacturer's instructions (Pishtaz Teb Diagnostics, Cat. no. PT-VNEX-100 Tehran, Iran). Target genes of SARS-CoV-2 were detected using an approved primer-probe-based Real-time PCR (Pishtaz Teb Diagnostics, cat. number. PT-COVID.19-100 Tehran, Iran). The fluorescence signal of HEX, FAM, and ROX was detected for the nucleocapsid gene and RdRP region of SARS-CoV-2, and RNase P served as the internal control. This detection was carried out using Rotor-Gene Q MDx 5plex HRM (Qiagen, Germany). A typical S-type amplification curve detected by the FAM or HEX channel, with Ct ≤ 40, indicated a positive SARS-CoV-2 virus.
3.3. Detection of Anti-SARS-CoV-2 IgM and IgG Antibodies
The serum-specific antibodies for SARS-CoV-2 (IgM and IgG) were tested using enzyme-linked immunosorbent assay (ELISA) kits following the manufacturer's instructions (Pishtaz Teb Diagnostics, Tehran, Iran). The IgM ELISA kit employed the IgM-captured sandwich method to detect the virus's nucleocapsid and spike proteins. IgG specific to nucleocapsid was detected through an indirect ELISA. In brief, for IgM detection, serum, and sample diluent were mixed in the appropriate wells coated with anti-human IgM antibodies and incubated for 30 minutes at 37°C. After washing, conjugates (nucleocapsid-HRP and spike-HRP) were added to the wells. For IgG detection, diluted serum specimens were applied to distinct wells coated with nucleocapsid and incubated for 30 minutes at 37°C. After discarding, conjugate (anti-human IgG-HRP) was added to the wells. In both IgM and IgG assays, after washing, chromogenic substrate was added to the wells and incubated for 15 minutes. The reaction was then stopped, and the absorbance of the wells was measured at 450 nm and 630 nm as the reference filter using an ELISA reader (BioTek Instrument Inc., Winooski, VT, USA). Samples with OD450/OD630 nm values above the cut-off value 1.1 were considered positive.
3.4. Measurement of Anti-SARS-CoV-2 Spike and RBD IgG Antibodies
The antibodies to spike and RBD in the serum of subjects were tested using indirect ELISA kits (Pishtaz Teb Diagnostics). Recombinant spike and RBD proteins were coated in the wells. The procedures were performed in the same manner as explained above for IgG to nucleocapsid. One hundred and two pre-pandemic normal serum samples collected 2 years before the COVID-19 pandemic were used to define the cut-off values (mean + SD). In this regard, 8 Ru/mL and 5 Ru/mL were considered as the cut-off values for anti-spike and anti-RBD IgGs, respectively.
3.5. SARS-CoV-2 Surrogate Neutralizing Antibodies (NAs) ELISA
NAs to SARS-CoV-2 were also measured using a functional surrogate NAs ELISA designed based on the RBD and human ACE2 interaction in a 96-well plate (Pishtaz Teb Diagnostics; cat. numbers: PT-CoV2NT-96). In this assay, serum and ACE2-HRP were mixed in the appropriate wells coated with RBD protein and then incubated for 30 minutes at 37°C. After adding chromogenic substrate into the wells, the absorbance of the developed colors was measured as described above. The quantity of NAs (µg/mL) was calculated based on the standard curve. The cut-off value for neutralizing antibodies was 2.5 µg/mL.
3.6. Statistical Analysis
Data were analyzed using SPSS v. 22.0 software (SPSS Inc., Chicago, IL, USA) and GraphPad Prism software version 8.0 (GraphPad Software Inc., San Diego, USA). Continuous variables were presented as mean ± SD, and categorical variables were reported as counts and percentages. The correlation of parameters was analyzed using the non-parametric Spearman's Rank-Order Correlation.
4. Results
4.1. The Characterization of COVID-19 Infected Patients
A total of 134 patients were included in this study, with 63 of them being female (47.0%). The median age was 54 ± 1.33 years for both sexes. Among these patients, 102 (76.1%) were confirmed COVID-19 cases, while 32 (23.9%) were suspected patients exhibiting symptoms of acute respiratory infection syndromes and/or abnormalities in chest CT images but tested negative for SARS-CoV-2 nucleic acid. The most frequently reported symptoms in COVID-19 patients were fatigue (122 out of 134; 91%), myalgia (98 out of 134; 73.1%), dyspnea (94 out of 134; 70.1%), headache (88 out of 134; 65.7%), and fever (83 out of 134; 61.9%) (see Appendix 1 in the Supplementary File). Among the patients, 1.5% (2 out of 126) were mild cases, 27.6% (37 out of 126) were moderate cases, 42.5% (57 out of 126) were severe cases, and 22.4% (30 out of 126) were critical cases. A total of 11 out of 134 patients (8.2%) required ICU-level care. Additional patient characteristics can be found in Appendix 4 in the Supplementary File.
4.2. Seroconversion of Antibodies Against SARS-CoV-2 in COVID-19 Patients
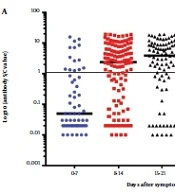
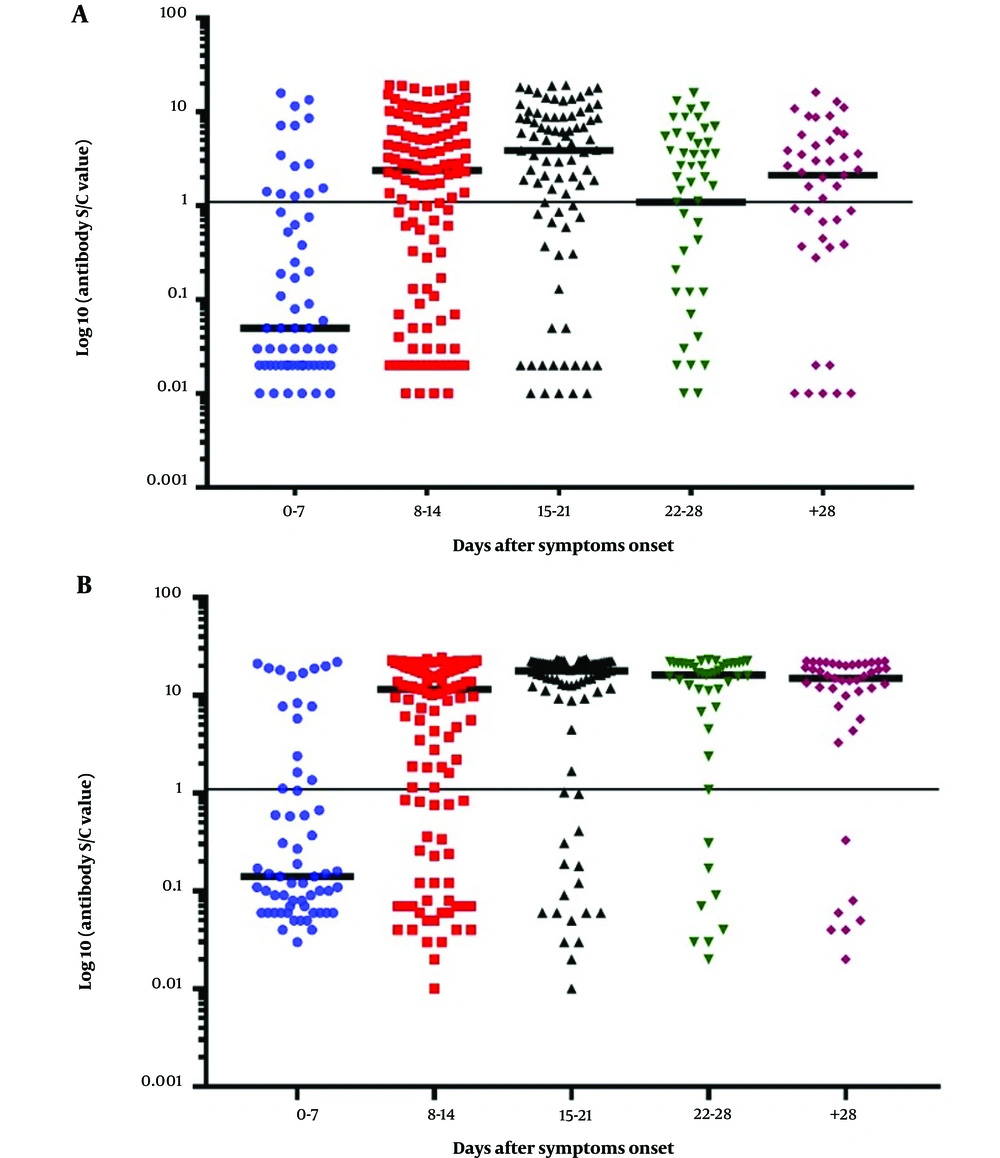
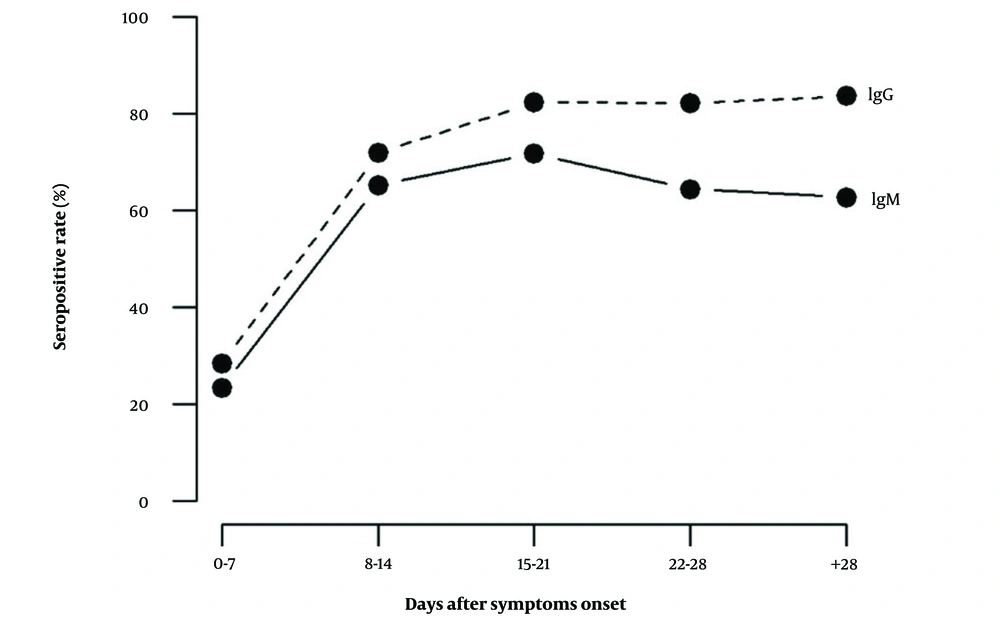
The median seroconversion time for IgM was 12 days, while for IgG, it was 10 days after the onset of symptoms. The proportion of patients testing positive for IgM was 23.3% within the first 7 days and increased to 71.8% between days 15 and 21 after symptom onset. Subsequently, IgM positivity gradually declined to 64.4% and 62.8% at 22 - 28 days and over 28 days after symptom onset, respectively (refer to Figure 1A). The proportion of IgG-positive patients was 28.3% within the first 7 days and 82.2% between days 22 and 28 after symptom onset (see Figure 1B). Following the 28th day since symptom onset, the percentage of positive IgG cases remained relatively consistent (see Figure 2).
Titers of SARS-CoV-2 Specific IgM and IgG Antibodies in Hospitalized COVID-19 Patients at Various Time Points. The sample/cut-off index is represented in dot plots for IgM (A) and IgG (B) in each serum sample. The cut-off values were calculated as sample optical densities (ODs) divided by the cut-off index, as specified by the manufacturer. The horizontal line indicates the cut-off value distinguishing between IgM and IgG positive and negative samples. The means of the cut-off value for each group, specific to SARS-CoV-2 IgM and IgG, are shown by the solid line.
Serological profile of anti-SARS-CoV-2 specific antibodies in hospitalized COVID-19 patients at different time points since onset of symptoms. The proportion of patients testing positive for IgM increased to 71.8% at 15 - 21 days after symptom onset. Following this period, IgM positivity gradually decreased. Meanwhile, the % of patients tested positive for IgG was 82.4% at 15 - 21 days after symptom onset, and the percentage of IgG-positive cases remained relatively consistent after that.
4.3. Evaluation of Antibody Profile Based on RT-PCR Findings in COVID-19 Patients
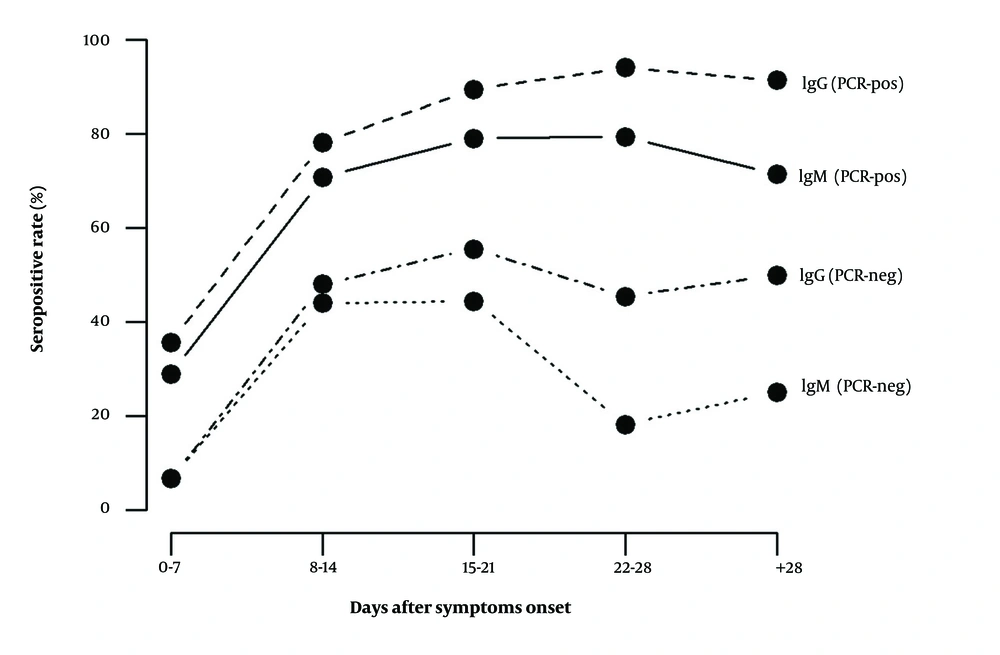
PCR testing at different time points revealed that the frequency of positive cases gradually decreased from 76.1% at the time of admission to zero at 36 days after sampling (see Table 1). Notably, SARS-CoV-2 was detected in 22% (11/50) of cases on day 14 and 9.1% (4/44) of cases on day 21 after sampling. Within the first 7 days of symptom onset, PCR testing showed the highest sensitivity at 76.1%, whereas the seroconversion rates for IgM (n = 85) and IgG (n = 90) were 23.3% and 28.3%, respectively. As illustrated in Figure 3, IgM and IgG seropositive patients had higher percentages in the PCR-positive group at all intervals. In the PCR-positive group, the seroconversion rates for specific IgM and IgG were 28.8% and 35.5%, respectively, one week after symptom onset. In PCR-negative patients, specific IgM and IgG were detected in only 6.7% (1/15) of cases one week after symptom onset (see Figure 3).
| Sampling Times (Days After Admission) | PCR Positive (%) | PCR Negative (%) |
|---|---|---|
| 0 | 102 (76.1) | 32 (23.9) |
| 3 | 43 (42.2) | 59 (57.8) |
| 7 | 2 (15.4) | 11 (84.6) |
| 14 | 11 (22) | 39 (78) |
| 21 | 4 (9.1) | 40 (90.9) |
| 36 | 0 (0) | 35 (100) |
SARS-CoV-2 Specific Real-time PCR Results in COVID-19 Patients at Different Time
Patterns of seropositivity for anti-SARS-CoV-2 specific IgM and IgG in RT-PCR positive and negative subgroups of hospitalized COVID-19 patients. At all intervals, the percentages of patients seropositive for IgM and IgG were higher in the PCR-positive group. In this group, the seroconversion rates for specific IgM and IgG were 28.8% and 35.5%, respectively, one week after symptom onset. In PCR-negative patients, specific IgM and IgG were detected in only 6.7% of cases one week after symptom onset.
4.4. Measurement of Anti-SARS-CoV-2 Spike and RBD IgG Antibodies
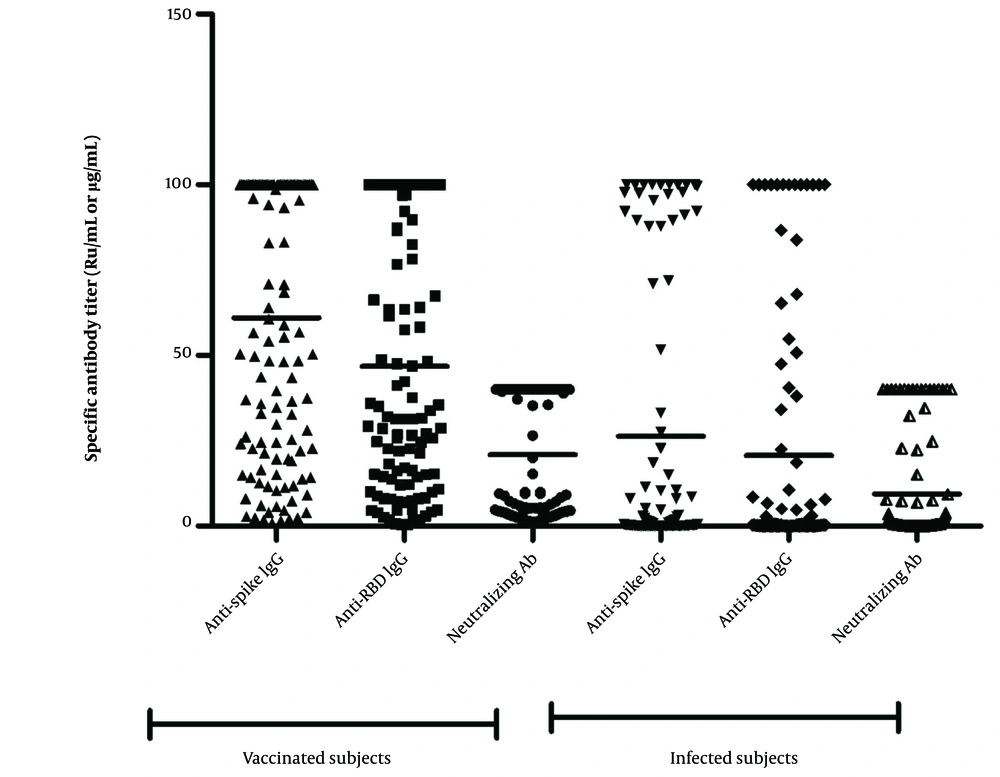
In vaccinated samples, 89.7% (122/136) tested positive for anti-spike IgG with a mean of 61.37 Ru/mL (95% CI: 54.86 - 67.87). Anti-RBD IgG was also detected in 87.39% (104/119) of vaccine recipients, with a mean of 46.86 Ru/mL (95% CI: 40.01 - 53.70). Among COVID-19 patients, 37.5% tested seropositive for anti-spike IgG (mean: 26.3 Ru/mL; 95% CI: 18.14 - 34.46), and 32.3% tested positive for anti-RBD IgG (mean: 20.6 Ru/mL; 95% CI: 13.30 - 28.04) (see Figure 4). Vaccine recipients had higher antibody titers than COVID-19 patients for both anti-spike and anti-RBD IgGs.
Levels of anti-spike and anti-RBD IgG and neutralizing antibodies (NAs) in COVID-19 vaccinated and infected subjects. The mean levels of anti-spike and anti-RBD IgGs and NAs were significantly lower in infected patients than vaccinated individuals. Therefore, anti-SARS-CoV-2 IgGs were produced more effectively in healthy vaccinees than the natural immunity to SARS-CoV-2 antigens in infected patients. The measurement units for anti-spike IgG, anti-RBD IgG, and neutralizing antibody are RU/mL, RU/mL, and µg/mL, respectively.
There was a significant correlation between anti-spike IgG and anti-RBD IgG concentrations in both COVID-19-infected individuals (P < 0.0001, r = 0.801) and vaccine recipients (P < 0.0001, r = 0.827). Antibodies to the RBD or spike protein were not detected in 7.1% of vaccinated samples, among which 7 individuals had received one dose of the Sputnik-V vaccine, and 3 had received two doses of the Sinopharm vaccine. Furthermore, the mean concentration of anti-spike IgG was higher in vaccine recipients with a history of COVID-19 (75.13 RU/mL) compared to those without previous infection (40.07 RU/mL) (P < 0.0001). A similar difference was observed for anti-RBD IgG, with mean concentrations of 56.93 RU/mL in vaccinees with a prior COVID-19 infection and 25.65 RU/mL in those without (P < 0.0001).
4.5. SARS-CoV-2 Surrogate Neutralization Antibody
In COVID-19 infected patients, NAs were detected in 32.3% (mean: 9.48 µg/mL; 95% CI: 6.32 - 12.63). Conversely, analysis of vaccine recipients showed that 87.9% produced NAs after vaccination, with a mean of 21.22 µg/mL (95% CI: 18.28 - 24.16) (see Figure 4). One out of 17 samples that tested negative for NAs had a history of COVID-19. Among the seronegative samples for NAs, nine out of 17 also tested negative for anti-RBD and anti-spike IgGs, and none of them had a history of COVID-19. There were significant correlations between anti-SARS-CoV-2 NAs and anti-spike IgG (r = 0.916) or anti-RBD IgG (r = 0.907) (P < 0.0001). Similarly, anti-SARS-CoV-2 NAs showed significant correlations with anti-spike IgG (r = 0.823) and anti-RBD IgG (r = 0.814) in COVID-19 infected patients. Consistent with anti-spike and anti-RBD IgG antibodies, the titer of NAs was higher in vaccine recipients with a history of COVID-19 (mean: 35.95 µg/mL) compared to those with no previous infection (mean: 13.46 µg/mL). Furthermore, among vaccinated subgroups, 15.0% (9/60), 6.1% (4/66), and 37.5% (3/8) were seronegative for NAs after receiving the first dose of Sputnik-V, 2 doses of Sinopharm, and 2 doses of COVXIN, respectively.
5. Discussion
The primary objective of this current cohort study was to investigate the antibody profiles of nucleocapsid, spike, and RBD proteins, as well as SARS-CoV-2 neutralizing antibodies (NAs) in Iranian COVID-19 patients and vaccine recipients. The kinetics of serum antibodies in COVID-19 patients indicated that the IgM titer increased until 15 - 21 days after symptom onset and then started to decline. Conversely, the IgG titer increased during the 15 - 21 day interval and remained stable until the last time point. Overall, the observed profiles of IgM and IgG antibodies align with findings from other studies (14, 15).
Additionally, the kinetics of anti-SARS-CoV-2 IgM and IgG responses revealed that seroconversion of IgG occurred prior to IgM in 11.9% of patients. Furthermore, 30.59% of COVID-19 patients simultaneously developed IgG and IgM antibodies at the first sampling time after symptom onset. In contrast to our results, several studies have reported synchronous seroconversion profiles of anti-SARS-CoV-2 IgG and IgM, or that IgM seroconversion occurs relatively earlier (6, 16-19). However, in line with our findings, some studies have reported earlier IgG responses than IgM (14, 19, 20). This discrepancy may be due to cross-immunity to SARS-CoV-2 (21), possibly arising from amino acid sequence homology between SARS-CoV-2 spike and nucleocapsid proteins and those of other Coronaviridae members (19, 22).
The RT-PCR test exhibited the highest sensitivity in hospitalized COVID-19 patients within the first 7 days of symptom onset, with a sensitivity of 76.1%. In contrast, seroconversion was positive in only 23.3% and 28.3% of patient sera for IgM and IgG, respectively, during the same time frame. Consistent with other studies (6, 23, 24), as the disease progressed, the sensitivity of serological tests increased, while the sensitivity of RT-PCR gradually declined to zero on the 36th day after sampling. This inconsistency is attributed to the initial lack of expansion of antibody responses in the early stages of infection (23). Our recent cross-sectional study on COVID-19 patients also demonstrated that the sensitivity of serological assays improved over time, with IgM and IgG test sensitivities increasing from 28.0% and 34.0% within 7 days of symptom onset to 51.3% and 61.5% beyond 7 days, respectively (25).
Given the varying sensitivity of these two tests at different stages of the disease, combining serological and molecular assays could enhance the diagnosis of SARS-CoV-2, particularly in RT-PCR negative cases. In line with our findings, the percentage of IgM and IgG seropositive patients was higher in RT-PCR-confirmed cases at all time intervals, with serum titers peaking at 15-21 days after symptom onset (26-28). Among RT-PCR-negative patients, who were admitted based on CT imaging and clinical signs and symptoms, SARS-CoV-2-specific IgM and IgG were detected in only 6.7% (1/15) of cases one-week post-symptom onset. Due to their negative molecular and serological results, we could not classify these individuals as COVID-19 cases, highlighting the importance of serological assays for more accurate assessments of COVID-19 cases (29). Additionally, antibody detection could be used alongside RT-PCR testing for diagnosing cases with negative RT-PCR results (19). In our study, we also assessed the titers of anti-spike and anti-RBD IgGs in both vaccinees and COVID-19 infected patients. A limitation of our study is the absence of anti-spike IgG and anti-RBD IgG testing in all cohort samples, which would have provided a clearer picture of antibody profile changes, enabling a more effective comparison between the COVID-19 infected and vaccinated groups. The mean levels of anti-spike and anti-RBD IgGs and NAs were significantly lower in infected patients compared to vaccines. Therefore, anti-SARS-CoV-2 IgGs were produced more efficiently in healthy vaccinees than through natural immunity to SARS-CoV-2 antigens in infected patients. This difference might be explained by the disparity in innate and adaptive immune cell activity in infected patients, impacting their ability to mount a humoral response to SARS-CoV-2. Lymphopenia and alterations in the functional commitment of T follicular helper cells (Tfh) (30) occur in COVID-19, potentially hindering the generation of an appropriate antibody response to SARS-CoV-2 antigens. However, the exact reasons for the difference in IgG response between infection and vaccination warrant further investigation.
Interestingly, mean concentrations of anti-spike and anti-RBD IgGs were higher in vaccinated individuals with a history of prior COVID-19 infection compared to those without previous infection. This aligns with the findings of Angyal et al., who investigated T cell and anti-spike IgG antibody responses in previously infected and SARS-CoV-2-naive healthcare workers who received a single dose of the BNT162b2 vaccine. They found that one vaccine dose elicited higher anti-spike IgG titers in individuals with previous SARS-CoV-2 infection than in SARS-CoV-2-naive individuals (31). Additionally, Buonfrate et al. reported that the median anti-RBD IgG titer following the first dose of the BNT162b2 mRNA COVID-19 vaccine was significantly higher in healthcare workers with a confirmed previous SARS-CoV-2 infection compared to those without prior infection (32).
Neutralizing antibodies (NAs) can directly inhibit SARS-CoV-2 from entering target cells (33). Accordingly, producing functional NAs at effective levels is a crucial component of protective immunity. Consistent with anti-spike and anti-RBD IgG findings, NAs were more abundant in vaccine recipients compared to COVID-19 infected patients. Vaccine recipients with a history of COVID-19 generated NAs more effectively than those with no prior infection. This is consistent with other studies reporting lower NAs in individuals vaccinated with CoronaVac who were naive to SARS-CoV-2 compared to vaccine recipients with a history of COVID-19 (34, 35). Early studies on mRNA vaccines also suggested that a single dose may be sufficient for individuals with a history of COVID-19, as strong antibody responses were observed after the first dose.
These studies have demonstrated robust antibody responses to the first dose of the vaccine in individuals who have recovered from COVID-19 (36-39). Therefore, vaccination can enhance memory B-cell responses specific to SARS-CoV-2 that were generated during a COVID-19 infection. In this regard, 94.1% of seronegative vaccine recipients for neutralizing antibodies (NAs) had no prior COVID-19 infection. Furthermore, 9 out of 17 individuals seronegative for NAs were also seronegative for anti-RBD and anti-spike IgG; all of these individuals had no history of COVID-19 infection. It is important to note that the genetic diversity among individuals, particularly in different allelic versions of HLA genes and other genetic loci, may account for the observed variations in antibody and T-cell responses across different studies (40, 41). Neither anti-RBD nor anti-spike antibodies were detected in 7.1% of the vaccinated samples, among which 3 individuals had received two doses of the Sinopharm vaccine.
Additionally, within the vaccinated subgroups, 6.1% (4 out of 66) and 37.5% (3 out of 8) were seronegative for neutralizing antibodies (NAs) after receiving two doses of the Sinopharm and COVAXIN vaccines, respectively. This result may be attributed to the limited efficacy of inactivated virus vaccines in activating SARS-CoV-2-specific T-cell responses, which are crucial for generating IgG specific to spike and RBD proteins, as well as NAs (8). It is evident that none of the available vaccines for SARS-CoV-2 are 100% effective and do not provide immediate protection post-vaccination.
5.1. Conclusions
In conclusion, the antibody profiles of IgM and IgG to SARS-CoV-2 demonstrated that the passage of time after symptom onset improves the sensitivity of serological assays, enhancing the diagnosis of COVID-19. The pattern of IgM and IgG detection suggests that in most patients, IgM and IgG can be detected simultaneously. Vaccine recipients displayed higher titers of anti-spike and anti-RBD IgGs and NAs compared to COVID-19 patients. Most seronegative vaccine recipients for NAs had no previous COVID-19 infection. Thus, vaccination appears to boost the immune response to prior SARS-CoV-2 infection.