1. Background
Cystic Echinococcosis (CE), a life-threatening zoonotic infection, is caused by the metacestode, a larval life stage of a dog tapeworm, of Echinococcus granulosus. Currently, ten genotypes (G1-G10) of genus Echinococcus sensu lato have been characterized by using mitochondrial markers of 16 species and 13 subspecies (1-3). Interestingly, the following taxonomic positions of these genotypes are defined based on an extensive revision: E. granulosus sensu stricto (G1 - G3), E. equines (G4) that are classified as non-zoonotic strains, and E. ortleppi (G5), E. canadensis (G6-G10) and E. felidis (‘lion strain’) as zoonotic strains (4-7). However, some genotypes have remained unknown in the consequence of overlapping cycles among various intermediate - definitive hosts (8, 9).
Epidemiologically, CE is distributed in hyperendemic regions of the world, including the Middle East, Northern and Eastern Africa, Central Asia, Mediterranean, Eastern Europe, and some foci in the United Kingdom (3, 10) where different intermediate hosts are circulating sympatrically. Nevertheless, due to the cross transmissions of Echinococcus species among diverse hosts and host-parasite interactions, the occurrence of mixed (dual/triple) infections would not be unexpected. In biological sequence analysis, based on bioinformatics tools, the mixed infections are suspected due to multiple overlapping peaks (chromatograms) observed in some sequences (11, 12). The lack of precise diagnosis of mixed infections leads to an insufficient understanding of the pathogenesis, classification, and biology of the parasite in pertinent regions.
To date, characterization of the mixed (concomitant) infections has been practically assisted by cloning and/or multiplex Real-Time PCR strategies that employ specific probes-primers in some parasitic infections (12-14). Some of the challenges of implementing these methods are their high cost and the need for reference laboratories, as well as designing specific probe-primers and optimizing conditions. Notably, discrimination of these infections has not been supported by PCR-RFLP and Nested-PCR methods due to the formation of nonspecific digestions and unidentified bands (15-17). Therefore, utilizing an available in silico program to analyze DNA sequences with heterozygous bases can reflect the valid status of neglected Echinococcus spp. in endemic areas. Furthermore, the identification of distinct genotypes by phylogenetic analysis together with clarifying their potentially novel strains/haplotypes demonstrates the differences in morphology aspects, transmission dynamics, and development and pathogenicity rate (1, 3, 7).
So far, at least four genotypes (G1, G2, G3, and G6) of E. granulosus have been reported in different parts of Iran (9, 18-20) where according to the circulation of a variety of intermediate hosts, this seems to be underestimated. However, many Iranian researchers are focusing on genotyping of E. granulosus in different regions of Iran and among populations of sheep, cattle, goat, and camel (19-24). With regard to the various intermediate hosts, including sheep, buffalo, pig, camel, goat, and cervid in these regions, the isolation and characterization of mixed genotypes as well as discrimination of their heterogeneity traits warrant a more realistic image of the parasite status in the region.
The present study aimed to introduce an in silico program for analyzing DNA sequences with heterozygous bases via phylogenetic analyses. This could facilitate the identification of E. granulosus isolates and provide insights into the determination of neglected Echinococcus spp. taxonomic status in central Iran.
2. Methods
2.1. Sample Preparation
A total of sixty hydatid cysts were obtained from hospitals and the slaughterhouse of Arak (Markazi province, geographically located in central Iran). The cysts were taken separately from 20 humans, 15 sheep, 15 goats, and 10 cattle liver and lung tissues. Then, these samples were aseptically fixed using 80% (v/v) ethanol. In addition, the presence of possible protoscolices was examined by optical microscopes and on aspirated hydatid fluids. The viability rate of protoscolices was confirmed by 0.1% aqueous eosin staining, preceded by washing with phosphate buffered saline (PBS, pH 7.2) (25). Whilst the live protoscolices remained colorless, dead ones absorbed eosin and became reddish. For additional molecular modalities, storage of the washed protoscolices was performed at 4°C in 70% ethanol.
2.2. Processes of DNA Extraction and Polymerase Chain Reaction (PCR)
Protoscolices taken from each sample were washed three times with phosphate-buffered saline (pH 7.2). Subsequently, they were sonicated to facilitate the breakdown of protoscolices. The extraction of total genomic DNA from either 50 μL of each samples’ protoscolices or germinal layers was performed based on the standard extraction procedure recommended by QIA DNeasy blood and tissue kit (QIAGEN, Hilden Germany) according to the manufacturer’s instructions. The extracted DNA concentration was assessed by NanoDrop system (thermo scientific Inc., Wilmington, DE). Furthermore, the extracted DNA was stored at -20°C until the molecular assessments. The forward (BD1:5’-GTC GTA ACA AGG TTT CCG TA-3’) and reverse primers (4S: 5’-TCT AGA TGC GTT CGA A(G/A)T GTC GAT G-3’) were used to amplify the 1000 bp ribosomal DNA (rDNA), also known as internal transcribed spacer 1 (ITS1). The following thermal cycling profile was exerted for each PCR reaction (total volume: 30 μL): 1 μL of genomic DNA (100 ng), 15 μL Master mix (2 ×), 1 μL (20 pmol) of primers (forward and reverse), and 12 μL Double distilled water (DDW). The primary denaturation cycle was conducted at 94°C for 5 minutes. 35 cycles of PCR contained secondary denaturation (45 seconds at 94°C), annealing (35 seconds at 55°C), primary extension (50 seconds at 72°C), and secondary extension (72°C for 7 minutes). DDW was used in all PCR assays representing the negative control. Also, 4 µL of PCR products was added to 1% agarose gel in the process of Electrophoresis, followed by ethidium bromide staining (45 minutes at 100 V). The bands were detected through a gel documentation system (KODAK gel logic 200 imaging system).
2.3. Restriction Fragment Length Polymorphism Polymerase Chain Reaction (PCR-RFLP) and Sequence Analysis
All PCR amplicons were digested for 2 - 3 hours at 37°C using restriction endonucleases RsaI and HpaII and buffers advised by the manufacturer (Fermentas, Vilnius, Lithuania). Then, ITS1-rDNA gene underwent endonuclease reaction in a volume of 30 µL. This reaction consisted of 1 µL of RsaI with cut site GT↓AC and 1 µL of HpaII (cut site C↓CGG) 10 µL of PCR products, 3 µL of 10× buffer, and 16 µL of distilled water. Electrophoresis of restriction fragments of amplicons was performed using a 2% (w/v) agarose gel at 100 V for 60 minutes. Indeed, the identification of Echinococcus spp. through RFLP method was determined based on the recently reported standardized pattern (9). Furthermore, direct sequencing of 21 amplicons by targeting ITS1-rDNA gene in both directions via the BD1 and 4S primers by ABIPRISMTM 3130 genetic analyzer automated sequencer led to the re-confirmation of RFLP results (applied biosystem, USA). The standard IUPAC codes used for combinations of two or more bases contributed to the coding of ambiguous sites. Contigs from all samples were aligned, justified, and edited in consensus positions compared to GenBank sequences of all regional species, attempting to determine the similarity of new haplotypes by employing the Sequencher Tmv.4.1.4 Software for PC (gene codes corporation) (http://multalin.toulouse.inra.fr/multalin).
2.4. Mixed Sequence Reader
The MSR program (freely available at http://MSR.cs.nthu.edu.tw/) has been developed not only to identify heterogeneity in the chromatographic traces, but also to establish the physical positions of the detected variants in the mixed infections. The algorithm used in MSR was modified from that of Indelligent, but MSR is designed to use reference database alignment. The analytic steps used in MSR were described by Chang et al. in 2012. In order to import the DNA sequence chromatographs, the imported files were chromatography traces in the ABI format. The base peak positions, quality values, and four channel signals (A, C, G, and T) recorded in the .abi format file were extracted and analyzed to identify the major and the minor signals at each base location.
2.5. Phylogenetic and Haplotype Network Assessments
A wide range of haplotypes based on the sequences of ITS1-rDNA using the statistical parsimony method was also applied by TCS 1.2 software (26). In addition, the possible limit of network estimation was 95%. Confidence limits with a 95% confidence interval were obtained for infection rates. A Neighbor-Net network built in splits tree 4.0 served to analyze the phylogenetic information provided by ITS1-rDNA sequences (27) regarding the genetic distances measured as well as the Kimura-2 parameter model of nucleotide substitutions. Finally, the heterogeneity of haplotype, Hd, and Nucleotide (π) was estimated by DnaSP version 5.10 software.
3. Results
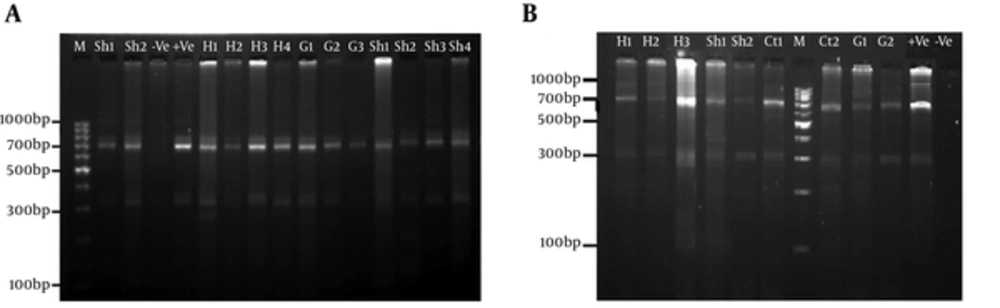
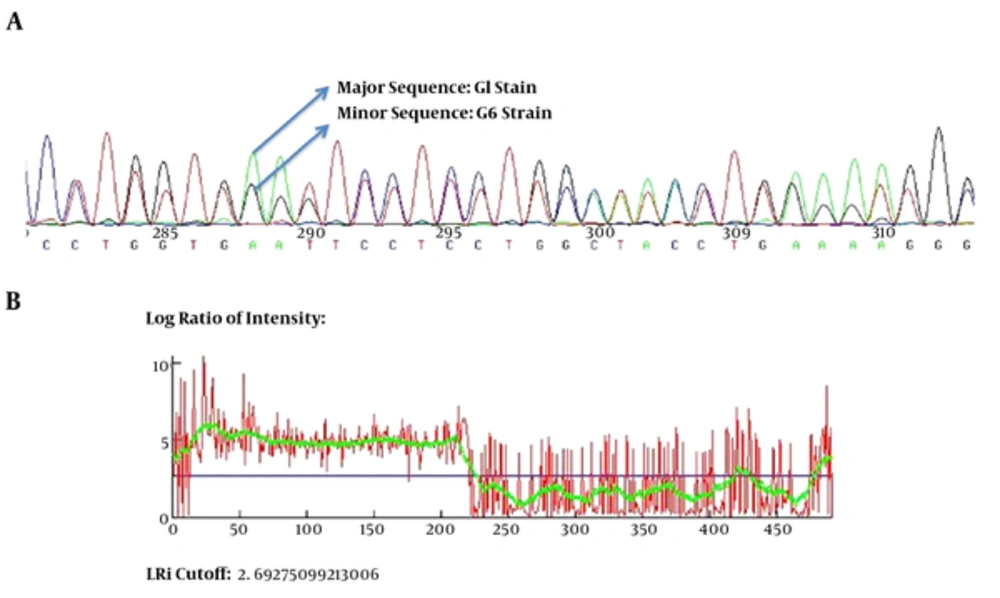
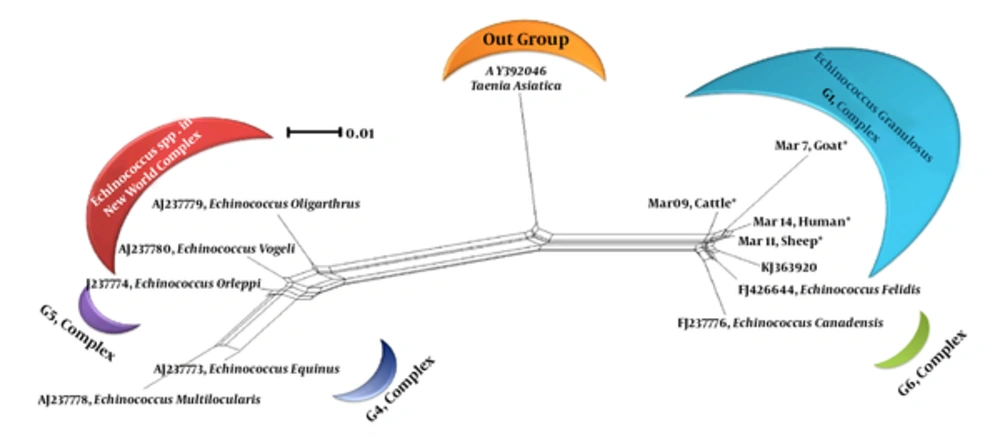
For all of the samples, a 1000 bp fragment was successfully amplified within ITS1-rDNA gene. Based on our in silico patterns, G1 (n = 58) was digested by restriction enzymes HpaII and RsaI into 300 bp, 700 bp and 345 bp, 655 bp fragments, respectively (Figure 1A and Figure 1B). However, no other expected genotype (such as G5) was distinguished in the cattle isolates. Furthermore, the digested fragments of mixed infections (n = 2) showed undefined patterns and were not supported by previously designed profiles (9). 21 out of 60 E. granulosus isolates (10 human, four sheep, four cattle, and three goat samples) were sequenced directly from ITS1-rDNA gene and determined firmly as corresponding to the 19 sheep strains (G1), two mixed infections. In addition, single-nucleotide variations (transition or transversion mutation models) were identified, including five unique haplotypes with a moderate range of diversity (Haplotype variability of 0.522 vs. Nucleotide variability of 0.02) in the constructed haplotype network. Two human (haplotype Mar 14), one sheep (haplotype Mar 11), one goat (haplotype Mar 07), and one cattle (haplotype Mar 09) isolates had seven nucleotide differences at positions 73, 74, 119, 160, and 312 bp. In addition, in an attempt to obtain the most likely major and minor variances, the BLAST analysis of overlapped chromatograms against a set of reference sequences showed that the isolated sequences of sheep strains belonged to infections with G1 and G6 genotypes (Figure 2A). The median mix ratio (major /minor) was 1.26. In addition, the LRi (log ratio of intensity) value for each sequence as the log ratio of the two intensities of the combined signal peaks was calculated with a cut-off of 2.69 (Figure 2B). Furthermore, the common haplotype Mar 08 (n = 16, frequency = 76%) was included without a notable heterogeneity in a consensus position. To identify a genealogical link within the haplotypes, we designed a statistical parsimony network. Figure 3 demonstrates all GenBank accession numbers for the inferred haplotypes of this study and for the reference genotypes/species used in the phylogenetic analysis.
4. Discussion
To date, one of the major difficulties in Echinococcus spp. taxonomic classification is related to the lake of accurate diagnosis of mixed infections in hyperendemic regions where different life cycles of Echinococcus spp. are sympatrically circulating with various heterogeneity traits and morphological aspects (21, 28). However, few studies have been carried out on the identification of Echinococcus spp. global concomitant infections (8, 29). In this study, an MSR program was used to directly analyze heterozygous base-calling chromatographs and subsequently detect the overlapped sequences and multiple structural variations, including SNPs (single nucleotide polymorphisms). The G1 and G6 were identified by MSR analysis showing that sheep and camel strains with novel haplotypes unequivocally exist in the region. These genotypes have been also reported in other central provinces of Iran (Semnan and Isfahan) (9); however, the type of mixed infections has not been determined precisely (30). Although G1 is the dominant strain in central Iran, the identification of both G1 and G6 genotypes in the region shows the presence of sheep-camel/dog life cycles in central Iran.
The liver and lung tissues of individual animals have shown the presence of the mixed infection caused by different haplotypes of E. granulosus spp. (8, 29). This can be arisen either from a single infection due to a definitive host concurrently harboring adult worms of the two haplotypes or genotypes, or from consecutive infections of the intermediate host. This study on 21 sequences of ITS1-rDNA gene led to the identification of merely five novel haplotypes in human, goat, cattle, and sheep isolates, while we expected to detect more haplotypes. In fact, this could be described by failing to use mitochondrial semi-conserved markers including cox 1 and nad 1. The ability of these markers in detecting more unknown haplotypes than nuclear-conserved gene (ITS1-rDNA) has already been proven (31, 32). The existence of novel haplotypes in G1 and mixed genotypes is related to the pathogenicity range of Echinococcus spp., the creation of re/emergent strains in the region, and the resistance of metacestodes to host innate immunity responses (33-35). Additionally, genotyping analyses of all cattle isolates exhibited the G1 strain. Nevertheless, we expected to detect other strains of E. granulosus such as E. ortoleppi since the cross-transmission of sheep and cattle genotypes of E. granulosus may occur in a wide range of hosts due to the overlapping cycles.
Several studies have attempted to genotype hydatid cysts in human, sheep, goat, cattle, buffalo, and camel populations in Iran on the basis of mitochondrial (cox1 and nad1) and nucleus markers, using RAPD-PCR, PCR-RFLP and sequencing strategies (19, 20, 22, 28, 30, 36, 37). However, none of them could provide remarkable applications in mixed genotypes. This issue authenticates the importance of discriminating overlapped E. granulosus genotypes existing in intermediate hosts to characterize the life cycle, host specificity, control programs, and classification of E. granulosus strains.
In summary, MSA, sequencing, and phylogenetic analyses showed that the common sheep strain (G1) and its concomitant infections with molecular diversity are unambiguously circulating in humans and livestock isolates in central Iran. The current results should be considered in improving the determination of the real epidemiological status of neglected Echinococcus spp. and launching disease control programs against echinococcosis previously understood life-cycle patterns. To appraise the multiple infection scenarios, further studies are needed focusing on cloning and multiplex real-time PCR in order to characterize the real taxonomic status of Echinococcus strains in wider areas of Iran and neighboring countries.