1. Background
Methicillin-resistant Staphylococcus aureus (MRSA) is considered a serious global health issue. Recent studies indicate an alarming increase in the incidence of MRSA over the past few years (1, 2). The annual mortality associated with MRSA infections (about 20,000 among hospitalized patients) is similar to the number of deaths due to AIDS, tuberculosis, and viral hepatitis in the United States (3). In Iran, the number of MRSA infections has extensively increased and MRSA is one of the most important multidrug-resistant organisms causing nosocomial infections (4, 5). Diabetes is one of the risk factors that increase susceptibility to MRSA colonization. The prevalence of this disease is predicted to rise from 382 million in 2013 to approximately 700.2 million in 2045. Approximately 15 - 25% of patients with diabetes may develop diabetic foot ulcers during their lifetime. Over 50% of these ulcerations can become infected, leading to high rates of hospitalization, lower limb amputation, and mortality (6, 7).
Methicillin resistance is due to the expression of a mutated penicillin-binding protein (PBP2a), with a low binding affinity for beta-lactams. This resistance mechanism allows cell wall biosynthesis to remain active even in the presence of typically inhibitory concentrations of almost all β-lactam antibiotics, including penicillinase-labile penicillins (e.g., penicillin G), penicillinase-stable penicillins (e.g., methicillin), and cephalosporins (e.g., cefoxitin) (8, 9). Penicillin-binding protein is encoded by the mecA gene, which is located on a mobile genetic element known as the Staphylococcal Chromosome Cassette mec (SCCmec) (10, 11). Typically, SCCmec has two essential components: The ccr gene complex, which is responsible for the mobility of the SCCmec cassette, and the mec gene complex (12).
To date, 15 different types of SCCmec have been described based on the combination of mec and ccr gene complexes (13). Analyzing epidemic MRSA clones using both phenotypic and genotypic markers is essential for developing effective strategies to control their spread, optimize antimicrobial therapies, reduce treatment-related costs, and understand the mode of pathogenicity (14). The phene-plate (PhP) system is a computerized biochemical fingerprinting method that analyzes the kinetics of bacterial biochemical processes in the presence of diverse metabolites. Compared to genotyping techniques such as pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST), the PhP system is specifically designed for each bacterial species to provide the highest discrimination among the strains of that species. The system is fast, easy to use, and provides valuable information for epidemiological studies of bacteria (15).
In addition to biochemical fingerprinting, SCCmec typing is one of the most important techniques routinely used to investigate the molecular epidemiology of MRSA clones (16). This method can distinguish between hospital-acquired MRSA (HA-MRSA) and community-acquired MRSA (CA-MRSA) strains (5, 17). Understanding the prevalence of MRSA infections among diabetic patients is crucial for treating and mitigating the spread of these pathogens. Therefore, there is an urgent need to study the prevalence and antimicrobial susceptibility patterns of MRSA isolated from patients with DFI.
2. Objectives
In the present study, we aimed to investigate the prevalence and diversity of clonal groups of MRSA strains isolated from patients with DFI in a major referral hospital in Tehran.
3. Methods
3.1. Isolation and Identification of MRSA Strains
Between April 2019 and March 2020, a total of 238 S. aureus isolates were collected from patients with DFI in a major referral hospital in Tehran, Iran. All isolates were cultured on HiCrome aureus agar medium (HiMedia, Mumbai, India), and single isolated colonies with a dark brown to black appearance were identified as S. aureus using polymerase chain reaction (PCR) with specific primers for the nucA gene (5). The isolates were saved in cryovials containing nutrient broth (Merck, Darmstadt, Germany) and 50% glycerol at -20ºC for further analysis.
3.2. Determination of Antibiotic Susceptibility Profiles
All confirmed S. aureus strains were tested for their susceptibility to cefoxitin, as a surrogate for methicillin, using the disk diffusion method based on the Clinical and Laboratory Standards Institute (CLSI) instructions (18). Identified MRSA isolates were further tested for their susceptibility against 17 antibiotics using the disk diffusion method. Antibiotic disks included penicillin (10 U), erythromycin (15 μg), clindamycin (2 μg), quinupristin-dalfopristin (15 μg), gentamicin (10 μg), amikacin (30 μg), kanamycin (30 μg), tobramycin (10 μg), nitrofurantoin (50 μg), ciprofloxacin (5 μg), tetracycline (30 μg), minocycline (30 μg), chloramphenicol (30 μg), linezolid (30 μg), rifampin (5 μg), trimethoprim-sulfamethoxazole (1.25 - 23.75 μg), and fusidic acid (10 μg). Additionally, the minimum inhibitory concentrations (MICs) of oxacillin and vancomycin were evaluated using the broth microdilution assay according to CLSI guidelines (18).
3.3. Phene-Plate Typing
All MRSA strains were typed using the PhP-CS plates (PhPlate AB, Sweden) containing four sets of dehydrated reagents, following the manufacturer's instructions as described previously (5). Microplates were incubated at 37°C, and the optical density (A620) was measured at 16, 40, and 64-hour intervals. The mean of three readings was calculated to produce a biochemical fingerprint for each isolate. Similarity among the isolates was calculated as a correlation (similarity) coefficient after a pairwise comparison of the biochemical fingerprints and clustered according to the unweighted pair group method (UPGMA) with arithmetic averages to yield a dendrogram. Isolates with the same fingerprint were regarded as belonging to the same PhP type. All data analysis, including the calculation of similarity among the isolates and the diversity of the bacterial populations, was performed using PhPWin software.
3.4. DNA Extraction
DNA of all isolates was extracted using the boiling method. In brief, a loopful of pure bacterial colonies was suspended in 300 µL of sterile distilled water and boiled for 20 minutes. After centrifugation at 13,000 × g for 15 minutes, the supernatant was used as the DNA template in the PCR reaction mixture (19).
3.5. Determination of SCCmec and ccr Types
All cefoxitin-resistant strains were confirmed as MRSA using specific primers for the mecA gene as described previously (5). The multiplex-PCR assay was employed to identify different types and subtypes of SCCmec (I, II, III, IVa, IVb, IVc, IVd, and V) as well as the type of ccr gene among MRSA strains (11, 13). The sequence of primers used for the identification and classification of SCCmec and ccr types is described in Appendix 1.
3.6. Detection of Panton-Valentine Leukocidin (pvl) Gene
The presence of the lukS/F-PV gene encoding the PVL S/F bicomponent protein was detected among MRSA strains by PCR reaction using specific primers (Appendix 1) (20).
4. Results
4.1. Identification of Strains
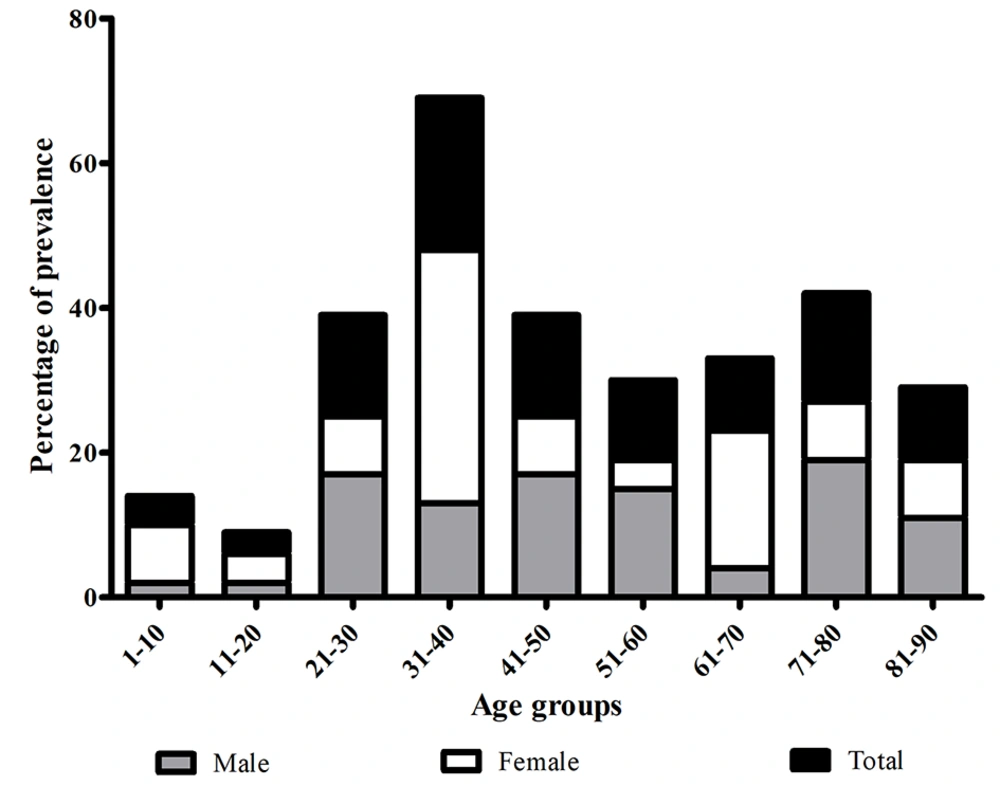
Of the 238 isolates confirmed as S. aureus, 73 (31%) were identified as MRSA based on their resistance to cefoxitin and the presence of the mecA gene. The highest number of DFI patients was observed among age groups between 31 - 40 years (n = 15, 21%) and between 71 - 80 years (n = 11, 15%), with most strains isolated from male patients in surgery and ICU settings (Figure 1 and Table 1).
| Sample | Intensive Care Unit | Surgery | Gynecology | Internal | Infectious Diseases | No. (%) |
|---|---|---|---|---|---|---|
| Male | 10 (21) | 19 (40) | - | 6 (13) | 12 (26) | 47 (64) |
| Female | 5 (19) | 8 (31) | 2 (8) | 4 (15) | 7 (27) | 26 (36) |
| Total | 15 (20) | 27 (37) | 2 (3) | 10 (14) | 19 (26) | 73 |
Distribution of MRSA Strains Isolated from Different Wards a
4.2. Antibiotic Susceptibility of MRSA Strains
Based on their antibiotic resistance patterns, MRSA strains were grouped into 29 antibiotypes (Table 2). All strains were resistant to penicillin, and 63 strains were resistant to more than six antibiotics, classifying them as multiple drug resistant. In contrast, all strains showed susceptibility to chloramphenicol, linezolid, quinupristin-dalfopristin, fusidic acid, and vancomycin and were therefore not included in Table 2. The highest level of antibiotic resistance was observed against ciprofloxacin (86%), followed by kanamycin (84%), tobramycin (84%), and erythromycin (82%). This study included ten MRSA strains from community patients, and they were all susceptible to the 13 antibiotics tested (except for penicillin) (Table 2).
| Pattern | Antibiotics | No. (%) of Strains | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD | TN | T | NI | K | AK | E | CIP | TS | RP | PG | GM | MN | ||
| 1 | + | 10 (14) | ||||||||||||
| 2 | + | + | + | + | + | + | + | + | 1 (1.4) | |||||
| 3 | + | + | + | + | + | + | + | + | + | 1 (1.4) | ||||
| 4 | + | + | + | + | + | + | + | + | + | + | 1 (1.4) | |||
| 5 | + | + | + | + | + | + | + | + | + | 4 (5.5) | ||||
| 6 | + | + | + | + | + | + | 1 (1.4) | |||||||
| 7 | + | + | + | + | + | + | + | 1 (1.4) | ||||||
| 8 | + | + | + | + | + | + | + | + | + | + | + | 1 (1.4) | ||
| 9 | + | + | + | + | + | + | + | + | + | + | + | 1 (1.4) | ||
| 10 | + | + | + | + | + | + | + | + | + | + | + | + | 2 (2.8) | |
| 11 | + | + | + | + | + | + | + | + | + | + | + | 6 (8.2) | ||
| 12 | + | + | + | + | + | + | + | + | + | + | 1 (1.4) | |||
| 13 | + | + | + | + | + | + | + | + | + | + | + | 3 (4.1) | ||
| 14 | + | + | + | + | + | + | + | + | + | + | + | 7 (9.6) | ||
| 15 | + | + | + | + | + | + | + | + | + | + | 2 (2.8) | |||
| 16 | + | + | + | + | + | + | + | + | + | + | 1 (1.4) | |||
| 17 | + | + | + | + | + | + | + | + | + | + | 6 (8.2) | |||
| 18 | + | + | + | + | + | + | + | + | + | + | 5 (6.8) | |||
| 19 | + | + | + | + | + | + | + | + | + | + | + | 3 (4.1) | ||
| 20 | + | + | + | + | + | + | + | + | 1 (1.4) | |||||
| 21 | + | + | + | + | + | + | + | + | + | + | 1 (1.4) | |||
| 22 | + | + | + | + | + | + | + | + | + | + | 1 (1.4) | |||
| 23 | + | + | + | + | + | + | + | + | + | + | 2 (2.8) | |||
| 24 | + | + | + | + | + | + | + | + | + | 1 (1.4) | ||||
| 25 | + | + | + | + | + | + | + | + | + | 2 (2.8) | ||||
| 26 | + | + | + | + | + | + | + | + | 5 (6.8) | |||||
| 27 | + | + | + | + | + | + | + | 1 (1.4) | ||||||
| 28 | + | + | + | + | + | + | + | + | 1 (1.4) | |||||
| 29 | + | + | + | + | + | + | 1 (1.4) | |||||||
Antibiotic Resistance Patterns of MRSA Strains
Based on the MIC values of oxacillin, 49% of MRSA strains were identified as high-level oxacillin-resistant (MIC ≥ 256 µg/mL), with only 5.5% of strains being highly susceptible (MIC = 4 µg/mL) to oxacillin. These strains all belonged to antibiotype 1. The MIC values of the strains toward vancomycin were generally low, ranging between 0.125 to 2 µg/mL (Table 3). While 60% of MRSA strains had a MIC below 0.5 µg/mL, 18% of strains showed resistance to 2 µg/mL of vancomycin.
| No | Gender | Ward | Year | PhP Type | Antibiotic Pattern | MIC OXA, µg/mL | MIC VAN, µg/mL | SCCmec Type | ccr Type | pvl | MRSA |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Male | ICU | 2019 | CT1 | 18 | 256 | 2 | III | 3 | - | HA- |
| 2 | Male | ICU | 2019 | CT1 | 4 | 96 | 1 | III | 3 | - | HA- |
| 3 | Female | Gynecology | 2019 | CT1 | 14 | 256 | 1 | III | 3 | - | HA- |
| 4 | Female | Infectious | 2019 | CT1 | 29 | 96 | 0.5 | III | 3 | - | HA- |
| 5 | Male | Surgery | 2019 | CT2 | 27 | 128 | 1 | III | 3 | - | HA |
| 6 | Male | Surgery | 2019 | ST1 | 20 | 256 | 0.25 | III | 3 | - | HA- |
| 7 | Female | Infectious | 2019 | CT3 | 14 | 256 | 2 | III | 3 | - | HA- |
| 8 | Male | ICU | 2019 | ST2 | 1 | 4 | 2 | IVa | 2 | + | CA- |
| 9 | Male | Gynecology | 2019 | CT2 | 17 | 96 | 1 | III | 3 | - | HA- |
| 10 | Female | Internal | 2019 | CT3 | 15 | 128 | 1 | III | 3 | - | HA- |
| 11 | Female | ICU | 2019 | CT2 | 17 | 256 | 0.5 | III | 3 | - | HA- |
| 12 | Female | Surgery | 2019 | CT2 | 22 | 256 | 0.5 | III | 3 | - | HA- |
| 13 | Male | ICU | 2019 | CT4 | 11 | 64 | 0.25 | III | 3 | - | HA- |
| 14 | Male | Internal | 2019 | CT2 | 23 | 256 | 0.125 | III | 3 | - | HA- |
| 15 | Male | Surgery | 2019 | CT2 | 28 | 256 | 0.25 | III | 3 | - | HA- |
| 16 | Female | Infectious | 2019 | CT2 | 24 | 64 | 0.125 | III | 3 | - | HA- |
| 17 | Male | Surgery | 2019 | CT3 | 12 | 256 | 0.5 | III | 3 | - | HA- |
| 18 | Female | Internal | 2019 | CT3 | 14 | 256 | 0.125 | III | 3 | - | HA- |
| 19 | Male | Infectious | 2019 | CT3 | 11 | 128 | 0.25 | III | 3 | - | HA- |
| 20 | Male | Surgery | 2019 | CT3 | 16 | 256 | 0.125 | III | 3 | - | HA- |
| 21 | Female | ICU | 2019 | ST3 | 1 | 4 | 0.5 | IVa | 2 | + | CA- |
| 22 | Male | ICU | 2019 | CT2 | 6 | 256 | 2 | III | 3 | - | HA- |
| 23 | Male | Surgery | 2019 | CT4 | 5 | 96 | 0.125 | III | 3 | - | HA- |
| 24 | Male | Surgery | 2019 | CT2 | 18 | 128 | 2 | III | 3 | - | HA- |
| 25 | Male | ICU | 2019 | ST4 | 1 | 8 | 0.125 | V | 5 | + | CA- |
| 26 | Male | Infectious | 2019 | ST5 | 1 | 4 | 0.5 | IVc | 2 | + | CA- |
| 27 | Female | Infectious | 2019 | ST6 | 1 | 6 | 1 | V | 5 | + | CA- |
| 28 | Female | Surgery | 2019 | CT2 | 26 | 256 | 1 | III | 3 | - | HA- |
| 29 | Female | Infectious | 2019 | CT4 | 1 | 8 | 2 | IVa | 2 | + | HA- |
| 30 | Male | Internal | 2019 | CT2 | 17 | 128 | 1 | III | 3 | - | HA- |
| 31 | Female | Surgery | 2019 | ST7 | 7 | 96 | 2 | III | 3 | - | HA- |
| 32 | Male | Surgery | 2019 | CT2 | 25 | 256 | 0.5 | III | 3 | - | HA- |
| 33 | Female | Infectious | 2019 | CT5 | 1 | 6 | 0.5 | IVa | 2 | + | HA- |
| 34 | Female | ICU | 2019 | CT2 | 23 | 256 | 1 | III | 3 | - | HA- |
| 35 | Male | Infectious | 2019 | CT2 | 26 | 256 | 0.5 | III | 3 | - | HA- |
| 36 | Male | Infectious | 2020 | CT2 | 25 | 64 | 0.5 | III | 3 | - | HA- |
| 37 | Female | Surgery | 2020 | CT2 | 26 | 96 | 1 | III | 3 | - | HA- |
| 38 | Male | Surgery | 2020 | CT3 | 14 | 256 | 0.125 | III | 3 | - | HA- |
| 39 | Male | ICU | 2020 | CT3 | 3 | 128 | 1 | III | 3 | - | HA- |
| 40 | Male | Surgery | 2020 | ST8 | 10 | 256 | 1 | III | 3 | - | HA- |
| 41 | Male | Infectious | 2020 | CT2 | 18 | 96 | 0.5 | III | 3 | - | HA- |
| 42 | Female | Surgery | 2020 | CT3 | 5 | 96 | 0.125 | III | 3 | - | HA- |
| 43 | Male | Infectious | 2020 | CT6 | 19 | 256 | 0.5 | III | 3 | - | HA- |
| 44 | Male | Surgery | 2020 | CT6 | 11 | 256 | 0.25 | III | 3 | - | HA- |
| 45 | Male | ICU | 2020 | CT3 | 14 | 256 | 0.125 | III | 3 | - | HA- |
| 46 | Male | Internal | 2020 | CT2 | 19 | 128 | 0.25 | III | 3 | - | HA- |
| 47 | Male | Infectious | 2020 | CT2 | 26 | 128 | 0.125 | III | 3 | - | HA- |
| 48 | Female | Infectious | 2020 | CT3 | 13 | 256 | 0.25 | III | 3 | - | HA- |
| 49 | Male | ICU | 2020 | CT2 | 10 | 96 | 0.25 | III | 3 | - | HA- |
| 50 | Male | Internal | 2020 | CT5 | 14 | 128 | 1 | III | 3 | - | HA- |
| 51 | Male | Surgery | 2020 | CT7 | 18 | 256 | 0.25 | III | 3 | - | HA- |
| 52 | Male | Infectious | 2020 | CT2 | 21 | 256 | 0.25 | III | 3 | - | HA- |
| 53 | Male | Surgery | 2020 | CT2 | 8 | 256 | 0.25 | III | 3 | - | HA- |
| 54 | Male | Surgery | 2020 | CT4 | 14 | 256 | 0.25 | III | 3 | - | HA- |
| 55 | Female | ICU | 2020 | CT7 | 13 | 256 | 0.25 | III | 3 | - | HA- |
| 56 | Female | Surgery | 2020 | ST9 | 1 | 8 | 0.25 | IVa | 2 | + | CA- |
| 57 | Male | Internal | 2020 | ST10 | 1 | 16 | 0.25 | V | 5 | + | CA- |
| 58 | Male | Surgery | 2020 | CT2 | 13 | 128 | 0.125 | III | 3 | - | HA- |
| 59 | Male | Infectious | 2020 | CT3 | 1 | 4 | 0.25 | IVc | 2 | + | CA- |
| 60 | Female | Surgery | 2020 | CT2 | 18 | 256 | 1 | III | 3 | - | HA- |
| 61 | Female | Infectious | 2020 | CT2 | 26 | 256 | 1 | III | 3 | - | HA- |
| 62 | Male | Surgery | 2020 | CT2 | 17 | 128 | 0.125 | III | 3 | - | HA- |
| 63 | Female | ICU | 2020 | ST11 | 15 | 256 | 2 | III | 3 | - | HA- |
| 64 | Male | Infectious | 2020 | CT2 | 17 | 128 | 0.125 | III | 3 | - | HA- |
| 65 | Male | Infectious | 2020 | CT8 | 5 | 256 | 2 | III | 3 | - | HA- |
| 66 | Male | Surgery | 2020 | CT5 | 11 | 256 | 0.25 | III | 3 | - | HA- |
| 67 | Female | Internal | 2020 | CT8 | 11 | 96 | 0.5 | III | 3 | - | HA- |
| 68 | Female | Internal | 2020 | CT8 | 5 | 256 | 1 | III | 3 | - | HA- |
| 69 | Male | ICU | 2020 | CT2 | 2 | 256 | 2 | III | 3 | - | HA- |
| 70 | Male | Internal | 2020 | CT2 | 17 | 128 | 0.25 | III | 3 | - | HA- |
| 71 | Female | Surgery | 2020 | CT4 | 11 | 256 | 2 | III | 3 | - | HA- |
| 72 | Male | Surgery | 2020 | CT2 | 9 | 256 | 2 | III | 3 | - | HA- |
| 73 | Male | Surgery | 2020 | CT2 | 19 | 128 | 2 | III | 3 | - | HA- |
Association Between PhP Types, SCCmec, ccr Types, and pvl Status (Positive or Negative) of the HA- and CA-MRSA Strains
4.3. Determination of SCCmec and ccr Types
In total, four different SCCmec types (III, IVa, IVc, and V) were detected among the MRSA strains, with 63 strains (86%) belonging to SCCmec type III (Table 3). Moreover, 14% of strains with SCCmec types IVa (7%), V (4%), and IVc (3%) carried the pvl gene and were classified as CA-MRSA strains. Additionally, ccr types 2, 3, and 5 were found among the strains. All CA-MRSA strains belonged to antibiotype 1 and showed low-level resistance to oxacillin (MICs < 16 µg/mL).
4.4. Clonality of MRSA Strains
PhP typing of the MRSA strains revealed the presence of diverse PhP types, consisting of 8 common types (CTs) and 11 single types (STs) (Appendix 2), with CT2 containing the highest number of MRSA strains (n = 31, 42%), followed by CT3 (n = 12, 16%) (Table 3). Except for CT6, all other CTs were common among both males and females. MRSA strains belonging to CTs 2, 3, 4, and 5 were commonly isolated from patients during 2019 and 2020, whereas strains belonging to CT1 were isolated only in 2019. All CA-MRSA strains belonged to CT3-CT5 and seven STs (i.e., ST2, ST3, ST4, ST5, ST6, ST9, and ST10).
5. Discussion
Diabetic foot infection (DFI) is a major health issue among patients with diabetes, and identifying the microbial strains causing these infections is crucial for determining the most suitable antibiotic therapy. MRSA strains have been reported as a primary problem in the treatment of DFI, particularly in the provision of appropriate empiric antimicrobial therapy (21). In this study, we aimed to determine the prevalence of MRSA among patients with diabetic foot ulcers in a referral hospital in Tehran, Iran. Interestingly, we found the prevalence of MRSA in DFI patients to be 4% higher than previously reported in the same region (21, 22). This gradual increase in infection rate might be due in part to the evolution of bacteria and/or the improper use of antibiotics in recent years (22).
Moreover, we observed almost twice the number of male patients with DFI and MRSA compared to female patients. This finding aligns with previous observations indicating a significant association between male gender and increased risk of MRSA acquisition. The higher attention to personal hygiene among women may contribute to this discrepancy, as noted by Ahmadi et al. (23). We also found a higher prevalence of MRSA infections among young adults (> 20 years) compared to pediatric and adolescent populations. This difference may be attributed to factors such as immune response variations in these age groups, as well as changes in nutritional status and anatomic and physiologic modifications (24).
The prevalence of MRSA infection also varied among patients hospitalized in different wards of the hospital, which could be attributed to variations in infection control practices and organizational factors in each ward. These findings align with previous studies reporting a higher rate of nosocomial infections in ICU and surgical wards compared to other units of the hospital (25, 26), suggesting a need for more stringent infection control measures in these wards.
In this study, all isolated MRSA strains were susceptible to chloramphenicol, linezolid, quinupristin-dalfopristin, and fusidic acid. The low levels of resistance to these drugs could be partly due to their unwanted side effects or the high expense of linezolid and quinupristin-dalfopristin in Iran (15, 19). In Iran, the selection of antibacterial drugs is made according to the ulcer depth and infection severity, based on global empiric antibiotic regimens for diabetic foot ulcers (22). On the other hand, ciprofloxacin and tobramycin are frequently prescribed for the treatment of moderate to severe cellulitis with ischemia (or significant local necrosis) and life- (or limb-) threatening infections, respectively (27). In our study, the highest level of resistance was found against ciprofloxacin, followed by tobramycin, suggesting an urgent need for an antibiotic with more efficacy in empirical therapy against DFIs.
Compared to hospital-acquired (HA)-MRSA strains, the level of resistance to different classes of antibiotics was lower among community-acquired (CA)-MRSA strains. Except for penicillin, all CA-MRSA strains were susceptible to all classes of antibiotics tested, although there were very low levels of resistance to oxacillin among them. Based on these findings, we suggest that CA-MRSA strains may rapidly convert to highly resistant bacteria and become the dominant source of MRSA infections in hospitals. This replacement may be attributable to the following reasons: First, the presence of smaller types of SCCmec (usually type IV and V) and fewer antimicrobial resistance genes, which remarkably decreases the fitness costs of antibiotic resistance in CA-MRSA strains; second, due to the low expression level of virulence factors, HA-MRSA strains can hardly cause infection in healthy people (28, 29).
In our study, only CA-MRSA strains harbored the pvl gene. PVL is a two-component pore-forming cytotoxin responsible for rapid necrosis, apoptosis, and destruction of leucocytes. This toxin can promote soft tissue and bone infections and is often associated with severe DFIs (30). Therefore, the implementation of effective preventative measures against infection and monitoring programs is necessary to reduce the spread of these strains in healthcare settings. Few studies have been conducted to describe the prevalence and dissemination of clonal groups of MRSA strains among patients with DFIs in Iran. In the present study, we used a combination of high-resolution PhP typing and SCCmec types, revealing the presence of diverse clonal groups of MRSA in DFI patients. CT2 was the predominant type, comprising more than 41% of strains, which is consistent with our previous findings in Iran (5, 13, 19, 31). Strains belonging to different clonal types had various antibiotic resistance profiles, indicating that members of these clones have disseminated and gained their antibiotic resistance independently.
One of the limitations of this study is that the data were obtained from a single-center study. Therefore, these results should be interpreted with care as they may not fully represent the prevalence and antimicrobial susceptibility profiles of MRSA strains among diabetes patients with foot ulcers in healthcare settings across the country. Furthermore, while the presence of the PVL gene among MRSA strains is of clinical interest, its detection alone does not necessarily enhance our understanding of the full virulence potential of the MRSA strains. Interestingly, in our study, PVL was found only in small numbers, all belonging to CA-MRSA strains, further supporting its low association with virulence potential or clinical outcomes associated with MRSA infections. Further studies are needed to better understand the role of PVL in the clinical outcomes of MRSA infections.
However, despite these limitations, this study is one of the few reports that include information about the origin, clonality, and antimicrobial resistance profiles of MRSA isolated from DFI in Iran. With the high number of patients with DFI attending clinical centers, MRSA strains can easily disseminate to the community. Combined with the uncontrolled use of antibiotics, they may gain further resistance and spread within the community. This calls for urgent preventative measures to control the spread of multidrug-resistant MRSA strains from hospitals to the community, as well as the adoption of appropriate therapeutic approaches for these patients.