1. Background
Fungal infections, particularly those caused by Candida species, have been on the rise worldwide, with Candida albicans, C. glabrata, C. krusei, C. parapsilosis, and C. tropicalis accounting for the majority of cases (1-5). Among these, C. albicans is the most prevalent species, harboring virulence factors such as biofilm formation and phenotypic diversity, which allow it to cause disease in humans (6). Although commonly present as part of the normal human microbiota, C. albicans can become pathogenic in immunocompromised or antibiotic-treated individuals, leading to invasive infections (7-9). Changes in the expression of its virulence factors enable C. albicans to transition from a commensal organism to an opportunistic pathogen (10-13).
Biofilm formation, hydrophobicity, ergosterol content, and secretory enzymes are key virulence factors that enhance the pathogenicity of C. albicans (14, 15). In particular, biofilm formation enables C. albicans to establish protective barriers against antifungal agents, contributing to drug resistance (16, 17). Hydrophobicity and biofilm production also facilitate the irreversible adhesion of the fungus to host tissues (18-20). Ergosterol, an essential component of fungal cell membranes, is a primary target for antifungal therapies (21).
Globally, C. albicans is responsible for a substantial proportion of invasive fungal infections, with an estimated 400,000 cases of candidiasis annually, particularly among hospitalized patients (22). However, the prevalence of C. albicans and the distribution of its virulence factors may vary by region, reflecting differences in healthcare practices, patient demographics, and environmental conditions (23). In Iran, candidiasis is an emerging concern, with recent studies indicating an increase in C. albicans infections, particularly in immunocompromised individuals (24-26). Hospital-acquired infections due to environmental contamination remain a significant issue in Iranian healthcare settings, highlighting the need for monitoring the virulence factors of hospital-associated C. albicans isolates.
2. Objectives
This study aimed to detect and compare the virulence factors of C. albicans from clinical and environmental samples in Iran, to identify potential cross-contamination routes between patients and their environment.
3. Methods
3.1. Sample Collection
This cross-sectional study, conducted between January 2020 and December 2021, involved the collection of 105 clinical samples (including urine, wound exudates, oral swabs, and vaginal swabs) and 165 environmental samples (from patient beds, drug trolleys, floors, nursing stations, sharp containers, high-touch surfaces, and portable equipment) suspected of containing C. albicans at Imam Khomeini Hospital in Ahvaz, Iran.
3.2. Sample Size Determination
The sample size for this study was determined based on standard guidelines for cross-sectional studies on C. albicans prevalence and virulence factors, as well as typical sample sizes used in similar research. Although a formal power analysis was not conducted, the chosen sample size of 270 (105 clinical and 165 environmental samples) was deemed adequate for descriptive statistical analysis and for comparing the prevalence and virulence of C. albicans between clinical and environmental sources. Previous studies on fungal infections and C. albicans prevalence in hospital settings typically involve sample sizes ranging from 100 to 300, depending on the diversity of the samples and the study's objectives (e.g., to assess species distribution, antifungal resistance, or virulence factor profiling).
In the absence of a formal sample size calculation, the total sample size for this study was considered sufficient to achieve the research objectives and allow meaningful analysis of the differences between clinical and environmental isolates. The sample collection period (24 months) provided ample opportunity to include all eligible samples that met the study’s inclusion criteria, ensuring a comprehensive assessment of C. albicans in the hospital environment.
Informed consent was obtained from all participants, and the study procedures adhered to the ethical principles outlined in the Declaration of Helsinki (27). The study was approved by the Ethics Committee of Ahvaz Jundishapur University of Medical Sciences (approval code: IR.AJUMS.MEDICINE.REC.1399.035).
3.3. Isolation of Candida albicans
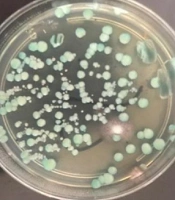
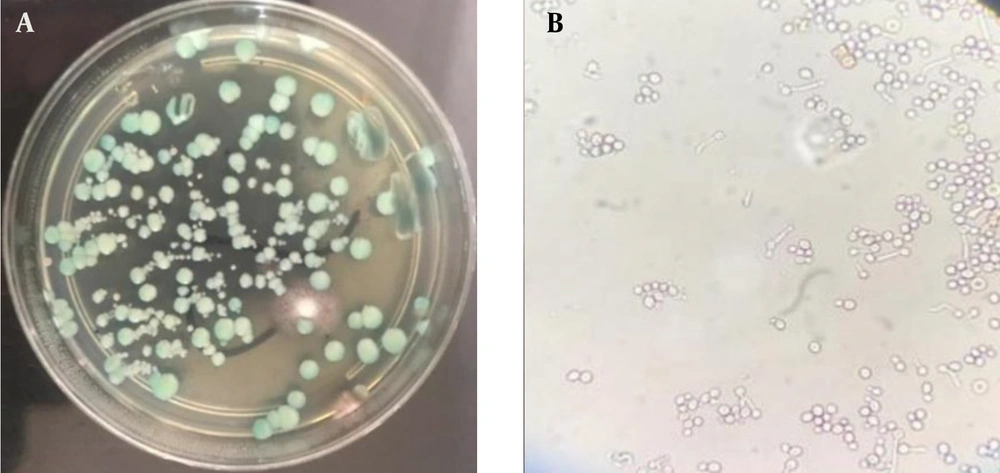
The clinical samples included 65 urine samples, 11 wound secretions, 10 oral cavity swabs, and 19 vaginal swabs. All clinical and environmental samples were cultured on CHROM agar medium (CHROM agar Candida, France) and incubated at 37°C for 37 - 48 hours. The growth of green colonies, suspected to be C. albicans, was assessed using phenotypic methods (28).
3.4. Phenotypic Identification
3.4.1. Germ-Tube Production
Suspected C. albicans isolates were cultured on Sabouraud dextrose agar (Conda, Spain) and incubated overnight at 37°C. A loopful of yeast colonies was added to 0.5 mL of fresh human serum and incubated for 2 hours at 37°C. The tube was gently shaken and examined under a light microscope at 40X magnification. Germ tube positivity was confirmed by the presence of a mass of long germ tubes branching from a yeast mother cell (29) (Figure 1).
3.5. Chlamydoconidia Formation Assay
The samples were cultured on Corn Meal agar (CMA) medium (Difco, USA) with 1% Tween 80 and incubated at room temperature for 3 - 5 days. Chlamydospore formation was then examined on the plates after incubation (29, 30).
3.6. Growth at 45ºC
To differentiate between C. dubliniensis and C. albicans, colonies were incubated at 45°C for 24 hours. C. albicans can grow at this temperature, while C. dubliniensis either does not grow or grows with difficulty, facilitating their differentiation (31).
3.7. Molecular Identification
To confirm the phenotypic tests, clinical and environmental isolates suspected of being C. albicans were selected, and their DNA was extracted using the boiling method described by Silva et al. (7). The pair of primers ITS1 (5'TCCGTAGGTGAACCTGCGG'3) and ITS4 (5'TCCTCCGCTTATTGATATGC'3) was used for molecular identification of C. albicans (8). Polymerase chain reaction (PCR) amplifications were carried out in a total volume of 25 µL. The PCR reaction contained 1 µL of lysed yeast cells, 12.5 µL of premix (Amplicon, Denmark), 0.2 µM of each primer (Integrated DNA Technologies), and sufficient deionized distilled water. The PCR program consisted of an initial denaturation step at 95°C for 5 min, followed by 35 cycles of denaturation at 95°C for 35 s, annealing at 58°C for 50 s, extension at 72°C for 1 min, and a final extension at 72°C for 10 min. Agarose gel electrophoresis (1.5%, Cinna Gen, Iran) was used to separate the PCR products, and C. albicans ATCC10231 was used as a positive control.
For restriction fragment length polymorphism (RFLP) analysis, 7 µL of PCR product was mixed with 1 µL of digest buffer, 0.5 µL of MSP1 restriction enzyme (Fermentas, Vilnius, Lithuania), and 5.7 µL of deionized distilled water. The mixture was incubated at 37°C for 24 hours. Then, 5 µL of the enzymatic reaction products were electrophoresed on a 2% agarose gel (Cinna Gen, Iran) for analysis.
3.8. Biofilm Formation Assay
To assess biofilm formation, isolated colonies were cultured on RPMI medium and incubated at 30°C for 24 hours in a shaking incubator. After 24 hours, all grown colonies were washed twice with sterile phosphate-buffered saline (PBS). Yeast suspensions were then prepared, and the concentration of all isolates was standardized using a spectrophotometer at 530 nm (Bio-Rad, USA) to an absorbance of 0.95 - 1.05. Twenty microliters of each suspension were added to 96-well flat-bottom plates, and 180 µL of RPMI was added to all wells. The microplates were incubated at 37°C for 48 hours. After incubation, the microplate contents were discarded, and the wells were washed twice with PBS to remove planktonic cells. Once dry, 100 µL of violet crystal dye was added to each well for 15 minutes. The dye was then discarded with distilled water, and excess stain was removed using 96% ethanol-acetone. The absorbance of each well was measured using an ELISA reader at a wavelength of 590 nm. The wells containing only 96% ethanol were used as a negative control. To evaluate biofilm formation, the mean absorbance of the negative controls was calculated and three times the standard deviation of the negative controls was added. Based on the cut-off value (ODc), the isolates were categorized into four groups: An absorbance less than or equal to ODc was considered non-biofilm producers; an absorbance greater than ODc but less than or equal to 2x ODc was considered weak biofilm producers; an absorbance greater than 2x ODc but less than or equal to 4x ODc was considered moderate biofilm producers; and an absorbance greater than 4x ODc was considered strong biofilm producers (9).
3.9. Ergosterol Content Assay
To prepare yeast isolates for analysis, an overnight culture of C. albicans was dissolved in 50 mL of Sabouraud dextrose broth (Biolife, Italy) and incubated at 35°C for 18 hours in a shaking incubator at 120 rpm. The suspensions were centrifuged at 2700 g for 5 minutes, and the sedimented yeast cells were transferred to a new 1.5 mL sterile microcentrifuge tube and incubated at -80°C for 24 hours. The samples were then lyophilized for 24 hours at -50°C under vacuum to dry the yeast isolates.
Next, 3 mL of alcoholic potash was placed in a sterile tube, and the dried yeast powder was added to it. The suspension was vortexed for 1 minute and placed in a bain-marie at 80°C for 1 hour. After cooling, 1 mL of distilled water and 3 mL of n-heptane were added to the tube, vortexed at high speed, and the supernatant (n-heptane layer) was transferred to a sterile tube and incubated at -20°C for 24 hours. Finally, the sterol content of the tubes was increased to 20 mL with 96% ethanol in a 1:5 ratio, and absorbance was measured using a spectrophotometer at wavelengths of 281.5 nm and 230 nm (10). The amount of sterol in the isolates was measured using the following equation.
Here, F is the dilution factor for the sample in ethanol, and 290 and 518 are the E values (in percentages per centimeter) determined for crystalline ergosterol and DHE, respectively.
3.10. Hydrophobicity Assay
To measure the cell surface hydrophobicity (CSH) of the isolates, isolated colonies were dissolved in Sabouraud dextrose broth and incubated for 24 hours at 25°C in a shaking incubator at 100 rpm. After 24 hours, all samples were centrifuged at 3000 g for 5 minutes. The supernatant was discarded, and each sample was washed twice with sterile PBS. Next, each sample was dissolved in a glass tube containing 10 mL of sterile distilled water, and 5 mL of the suspension was transferred to another sterile glass tube. The OD of the first series of tubes containing 5 mL of distilled water and yeast solution was read at 600 nm using a spectrophotometer, and this reading was recorded as OD0.
One mL of xylene was added to the second series of tubes containing 5 mL of distilled water and yeast suspension. The tubes were fully closed with paraffin and vortexed for 30 seconds. All tubes were then placed in a bain-marie at 30°C for 30 minutes. After 30 minutes, the xylene at the top of the tubes was discarded. The tubes were then boiled in a bain-marie at 100°C for 100 minutes. After cooling, the OD values were read at 600 nm and recorded as OD1 (11, 12).
The CSH of the isolates was calculated using the following equation.
This formula calculates the percentage of hydrophobicity based on the difference between the OD0 and OD1 values.
3.11. Extracellular Enzymes Assay
3.11.1. Phospholipase Activity
The phospholipase activity test medium was primarily composed of 65 g of Sabouraud dextrose agar, 5.5 g of calcium chloride, 58.4 g of NaCl, and supplemented with 2% egg yolk. To prepare the suspension of the overnight culture of C. albicans, sterile distilled water was used, and OD values were adjusted using a spectrophotometer at 640 nm to an absorbance range of 0.8 - 0.13.
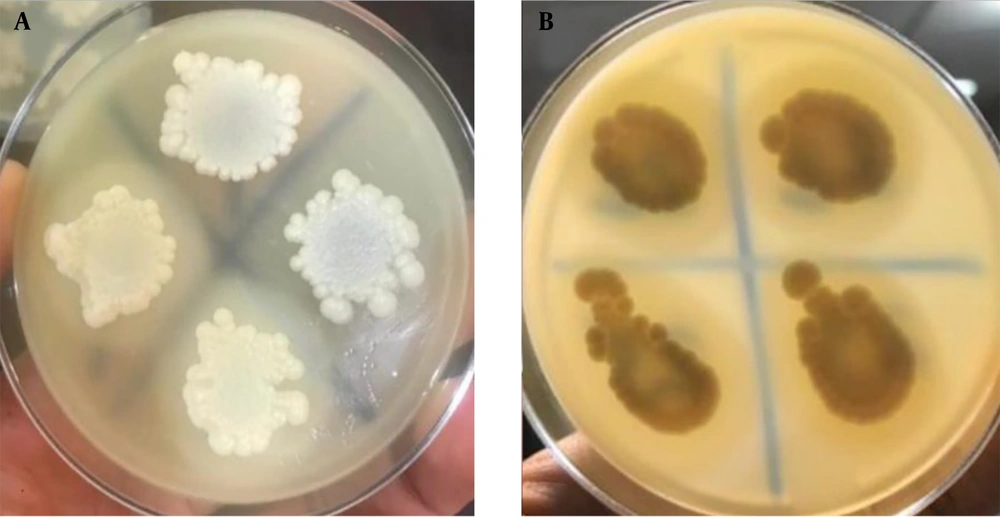
Five μL of each suspension were inoculated in triplicate onto the phospholipase medium and incubated at 37°C for 1 week (13). The zone of phospholipase activity inhibition for each strain was measured (14).
3.11.2. Proteinase Activity
To assess the proteinase activity of C. albicans isolates, bovine serum albumin agar (BSA) medium was used (15). The main components of BSA included 1 g of KH2PO4, 0.5 g of MgSO4, 10 g of glucose, 0.1 g of yeast extract, 2 g of bovine albumin, and 20 g of agar. All listed compounds, except for agar, were dissolved in 50 mL of distilled water, and the pH was adjusted to 5.
Overnight suspension cultures of isolates, equivalent to 0.5 McFarland standards, were prepared for each isolate. Five μL of each suspension were inoculated in triplicate onto the BSA medium and incubated at 37°C for 8 - 10 days. The zone of proteinase activity inhibition for each strain was measured.
3.12. Statistical Analysis
Statistical analysis was performed using SPSS v.18 (SPSS Inc., Chicago, IL, USA) and Stata version 14.0 (IBM, Armonk, NY, USA). Descriptive statistics, including frequencies and percentages for categorical variables, and means with standard deviations for continuous variables, were calculated. To compare categorical variables, such as phospholipase and proteinase activity and biofilm formation, the chi-square test was used. For continuous variables, such as hydrophobicity and ergosterol content, the independent t-test was applied to assess differences between clinical and environmental isolates.
The Kolmogorov-Smirnov test was used to assess the normality of the continuous data. In cases where data were not normally distributed (e.g., hydrophobicity), the non-parametric Mann-Whitney U test was applied. Statistical significance was set at a P-value of less than 0.05. All analyses were two-tailed.
4. Results
In this cross-sectional study conducted at Imam Khomeini Hospital in Ahvaz, Iran, a total of 105 clinical and 165 environmental samples were collected. The study included 86 patients, of whom 81.9% were women, aged between 20 and 70 years (Table 1). The majority of the clinical specimens (53.3%) were isolated from urine samples. Using both phenotypic and molecular methods, 30 (28.6%) clinical isolates and 30 (18.2%) environmental isolates were identified as C. albicans (Table 2) (Figures 1 and 2).
| Variabels | Candida albicans Positive | Candida albicans Negative |
|---|---|---|
| Samples | ||
| Urine | 16 (53.4) | 49 (65.4) |
| Wound secretion swabs | 7 (23.3) | 4 (5.3) |
| Oral cavity swabs | 1 (3.3) | 9 (12) |
| Vaginal swabs | 6 (20) | 13 (17.3) |
| Total | 100 | 100 |
| Gender | ||
| Male | 8 (26.7) | 15 (20) |
| Female | 22 (73.3) | 60 (80) |
| Total | 100 | 100 |
a Values are expressed as No. (%) unless otherwise indicated.
a Values are expressed as No. (%).
Out of the 30 C. albicans isolates identified, 12 (40.0%) clinical and 7 (23.3%) environmental isolates did not secrete the phospholipase enzyme. No statistically significant relationship was found (P = 0.262) between phospholipase enzyme secretion and the type of sample. Among the 60 C. albicans-positive isolates, 10 clinical (33.3%) and 7 environmental (23.3%) isolates did not exhibit proteinase activity. A statistically significant relationship was observed (P = 0.008) between proteinase enzyme secretion and the type of sample. The most prevalent C. albicans environmental isolates showed moderate phospholipase and proteinase activity (Figure 2) (Table 3).
| Samples | P-Value | |||
|---|---|---|---|---|
| Negative | 0.4 - 0.69 | 0.1 - 0.39 | ||
| Phospholipase | 0.262 | |||
| Clinical | 12 (40) | 11 (36.7) | 7 (23.3) | |
| Environmental | 7 (23.3) | 17 (56.7) | 6 (20.0) | |
| Proteinase | 0.008 | |||
| Clinical | 10 (33.3) | 10 (33.3) | 10 (33.3) | |
| Environmental | 7 (23.3) | 21 (70.0) | 2 (6.7) | |
a Values are expressed as No. (%).
Biofilm formation ability was detected in all 30 clinical isolates and 24 out of 30 environmental isolates. Clinical isolates were mostly strong biofilm producers, while environmental isolates exhibited weak biofilm formation ability. A statistically significant relationship (P < 0.001) was found between biofilm formation and the type of sample (Table 4).
| Samples | Biofilm Formation Scale | Total | P-Value | |||
|---|---|---|---|---|---|---|
| Negative | Weak | Moderate | Strong | |||
| Clinical | 0 (0) | 0 (0) | 3 (10) | 27 (90) | 30 (100) | < 0.001 |
| Environmental | 6 (20) | 21 (70) | 2 (6.7) | 1 (3.3) | 30 (100) | |
a Values are expressed as No. (%).
Cell surface hydrophobicity was higher in clinical isolates (66.4 ± 9.8) compared to environmental C. albicans isolates (47.7 ± 17.0). Ergosterol analysis showed no significant difference in content between clinical and environmental isolates (P = 0.517) (Tables 4 and 5).
| Samples | Number | Mean ± SD | P-Value |
|---|---|---|---|
| Hydrophobicity | < 0.001 | ||
| Clinical | 30 | 66.4 ± 9.8 | |
| Environmental | 30 | 47.7 ± 17.0 | |
| Ergosterol | 0.517 | ||
| Clinical | 30 | 1.2 ± 0.5 | |
| Environmental | 30 | 1.1 ± 0.3 |
5. Discussion
The prevalence of C. albicans among clinical specimens in this study was found to be 28.6%, which is somewhat lower than the 32 - 45% prevalence reported in previous studies (16-18). Interestingly, we observed a higher proportion of C. albicans in urine samples (53.4%) compared to other specimen types, which is consistent with previous reports (19, 20). Regarding the distribution among patients, the predominance of C. albicans in adult women aged 20 - 70 years (73.3%) reflects global trends, as this group is more susceptible to vaginal candidiasis (17, 20). Our recovery rate of C. albicans from the hospital environment (18.2%) is in close alignment with the results of Ferreira et al., who reported an 11% C. albicans isolation rate from the hospital environment (21).
Phenotypically, we confirmed the identity of C. albicans using standard methods, including chromogenic culture, germ tube formation, chlamydospore production, and growth at 45°C. These phenotypic tests have shown excellent specificity and sensitivity for C. albicans identification in previous studies (28, 32). Molecular confirmation of all isolates by PCR-RFLP in our study corroborates findings by other researchers who have advocated for the combination of phenotypic and genotypic analyses to accurately delineate C. albicans (29, 30).
In summary, the distribution of C. albicans among specimen types and patient groups, as well as the contamination rates and identity confirmation of isolates, are largely consistent with prior reports. Further comparisons of clinical and virulence attributes are warranted.
In this study, we measured the biofilm formation ability, ergosterol content, hydrophobicity, and phospholipase and proteinase activities of clinical and environmental isolates of C. albicans. The secreted hydrolytic enzymes phospholipase and proteinase are recognized as key mediators of C. albicans virulence and invasion (32, 33). In our study, while no significant difference in phospholipase activity was observed between clinical and environmental isolates (P = 0.262), proteinase activity was significantly higher in clinical isolates (P = 0.008). This suggests that proteinase may enhance the invasive capacity of clinical strains. We detected lower proteinase activity rates of 66.7% in clinical isolates and 76.7% in environmental isolates, compared to nearly universal production reported elsewhere (31, 33). Total phospholipase activity in clinical (60%) and environmental (76.7%) isolates was also lower than in previous studies (31, 34). These discrepancies likely reflect variations in methodology, geography, and strain types across different investigations. Indeed, our isolates exhibited a range of phospholipase and proteinase activities, highlighting the complex interplay between these virulence factors, isolates, and pathogenicity, which warrants further investigation. Nonetheless, the production of these hydrolytic enzymes by most isolates underscores their critical role in C. albicans virulence.
The findings are consistent with other studies reporting C. albicans' ability to form biofilms (35-37). A significant difference was found in biofilm-forming abilities between clinical and environmental isolates (P < 0.001). Most clinical isolates showed strong biofilm formation, while environmental isolates exhibited weak biofilm formation, in agreement with the findings of another study (38). This suggests that increased biofilm production likely influences C. albicans' pathogenicity.
Cell surface hydrophobicity of C. albicans is recognized as an essential factor in the yeast's adhesion to both abiotic and biotic surfaces, which may be involved in its virulence (35, 39). A significant difference was observed in CSH between clinical and environmental C. albicans isolates (P < 0.001). Clinical isolates exhibited higher hydrophobicity (66.4 ± 9.8) compared to environmental isolates (47.7 ± 17.0), consistent with the findings of Hazen et al. (36). This suggests a role for CSH in C. albicans' virulence.
Ergosterol is an essential sterol in the cell membranes of fungi, and changes in its biosynthetic pathway can be lethal to the fungal cell (37, 38). Ergosterol analysis showed no significant difference in content between clinical and environmental isolates (P = 0.517), highlighting its importance to both groups of isolates.
5.1. Limitations of the Study
Several limitations of this study should be noted. First, while our sample size of 270 isolates (105 clinical and 165 environmental) was sufficient for descriptive analysis, it may not fully capture the diversity of C. albicans strains encountered across a broader population or in different hospital settings. A larger, multicenter study would provide more robust data and allow for better generalizability of our findings.
Second, the cross-sectional nature of this study limits our ability to infer causality or assess the temporal dynamics of C. albicans transmission between clinical and environmental reservoirs. Longitudinal studies tracking the persistence of C. albicans strains over time in both clinical and hospital environments would offer valuable insights into transmission patterns and the factors influencing the development of hospital-acquired infections.
Third, while our study focused on the recovery and characterization of C. albicans isolates, it did not assess potential environmental factors such as hospital hygiene practices, cleaning protocols, or air quality, all of which could influence the presence and persistence of C. albicans in the hospital environment. Incorporating these factors into future studies could provide a more comprehensive understanding of the environmental conditions that facilitate fungal transmission and colonization.
Finally, while we assessed several key virulence factors—biofilm formation, proteinase and phospholipase activities, and hydrophobicity—other important virulence attributes, such as antifungal resistance mechanisms, adherence to host cells, and immune evasion strategies, were not included in this study. A more comprehensive profiling of C. albicans virulence factors, including genetic and transcriptomic analyses, would contribute to a deeper understanding of the mechanisms underlying its pathogenic potential.
5.2. Conclusions
This study provides a comprehensive characterization of C. albicans isolates from clinical and environmental samples in a hospital setting. Our findings demonstrate that the prevalence and distribution of C. albicans among clinical specimens align with those reported in previous studies, with a higher prevalence in urine samples and a predominance among adult women, consistent with known epidemiological patterns. Phenotypic and molecular analyses confirmed the identity of the isolates, reinforcing the importance of combining both approaches for accurate identification.
We identified significant differences between clinical and environmental isolates in several key virulence factors. Clinical isolates exhibited enhanced proteinase activity, stronger biofilm formation, and greater CSH compared to environmental isolates. These attributes suggest that clinical isolates possess a greater invasive potential, likely contributing to the pathogenicity of C. albicans in infections. In contrast, environmental isolates demonstrated attenuated virulence characteristics, although they still retained some pathogenic potential, such as phospholipase activity and biofilm formation.
Ergosterol content did not differ significantly between the two groups, highlighting its essential role across both clinical and environmental strains. Overall, our study underscores the complex interplay between virulence factors, isolate origin, and pathogenicity. Further research into the molecular mechanisms underlying these differences is necessary to better understand the transmission dynamics and pathogenic potential of C. albicans in hospital environments.