1. Background
Urinary tract infections (UTIs) are the second most common infections in humans after the common cold (1, 2). Urinary tract infections are a serious public health concern and can lead to severe, life-threatening conditions, primarily due to their recurring nature (3). Recurrent UTIs are directly related to biofilms formed within the urinary tract or on indwelling devices (4). Staphylococcus aureus is considered the second most prevalent etiologic agent in UTIs, causing persistent biofilm-related infections (2). In this bacterium, the main component of the biofilm matrix is polysaccharide intercellular adhesin (PIA), encoded by the intercellular adhesion operon (5). In addition to the ica operon, proteins belonging to the microbial surface components recognizing adhesive matrix molecules (MSCRAMMs), such as fibronectin-binding proteins A and B (fnbA and fnbB) and clumping factors A and B (clfA and clfB), mediate interactions between the bacterial cells and host extracellular matrix proteins. Additionally, the accessory gene regulator (agr) locus in S. aureus encodes the quorum sensing system, which plays a crucial role in regulating biofilm development (6, 7). Staphylococcus aureus isolates can be classified into four major agr groups (types I to IV) based on the sequences of agrC [encoding the receptor of the auto-inducing peptide (AIP)] and agrD (encoding a cyclic AIP) genes. Previous studies have emphasized that different agr types have varying effects on disease severity due to the relationship between the agr group and the genetic background of each strain (8).
Biofilm formation also facilitates the exchange and dissemination of genes involved in antibiotic resistance, resulting in limited treatment options, especially in infections caused by methicillin-resistant S. aureus strains (MRSA) (9, 10). Methicillin resistance is due to the presence of mecA and mecC genes, which are part of a large and mobile genetic element known as staphylococcal cassette chromosome mec (SCCmec). To date, 13 SCCmec types (according to the combination of ccr gene complexes and mec gene complexes) have been identified among MRSA strains (11). While vancomycin remains the primary treatment for MRSA infections, its high cost, intravenous administration, and several side effects limit its routine use. Although quinolone antibiotics have been proposed as an oral alternative to parenteral vancomycin therapy, the widespread use of these drugs in recent years has led to the emergence of fluoroquinolone-resistant S. aureus, especially among MRSA strains (9).
Aminoglycosides, one of the important drugs that enhance the activity of anti-staphylococcal antibiotics, are closely associated with methicillin resistance in S. aureus (10, 12). Therefore, understanding the frequency of genes involved in antibiotic resistance and analyzing epidemic S.aureus clones is essential to developing effective approaches for controlling the spread, improving antimicrobial therapies, and decreasing treatment-related costs (13). Among various phenotyping and genotyping methods, the phene plate (PhP) system and SCCmec typing are simple, fast, and inexpensive methods that provide beneficial information for epidemiologic studies of bacteria (14).
2. Objectives
This study aimed to determine the prevalence of clonal groups of S. aureus strains isolated from patients with UTIs in Isfahan, Iran. Additionally, we evaluated the possible genetic relationships between agr types, biofilm formation, and resistance to aminoglycosides and quinolones.
3. Methods
3.1. Sample Collection and Characterization of Staphylococcus aureus Isolates
In this study, a convenience sampling method was used for the collection of bacterial isolates. The sample size was calculated using the following formula based on the prevalence of biofilm formation among S. aureus strains (95.45%) reported in previous studies (15):
Where n = required sample size, p = prevalence of biofilm formation (95.45%), q = 1 – p, e = margin of error (5%), z = 2.58 at 99% CI.
A total of 119 suspected S. aureus isolates were obtained from patients with symptomatic UTIs at the laboratory of a referral hospital in Isfahan, Iran, in 2017. All patients exhibited symptoms of UTIs, such as frequent or urgent urination, dysuria, hematuria, fever, cloudy urine, tenderness, and suprapubic and flank pain. All bacterial isolates were cultured on nutrient agar (Biolife, Italy) medium (16). Pure colonies were collected and characterized as S. aureus strains using molecular tests.
3.2. Biofilm Formation Assay
3.2.1. Qualitative Method
Qualitative detection of biofilm production was performed using the Congo-red agar (CRA) method. The confirmed S. aureus strains were streaked on CRA plates (Merck, Germany). After 48 hours of incubation at 37°C and an additional overnight incubation at room temperature, the characteristics of each colony were examined to evaluate the potential for biofilm formation. Strains showing rough black colonies were interpreted as slime-positive (strong biofilm formation), those showing dark red colonies were considered suspected colonies (intermediate biofilm formation), and those showing light red colonies were interpreted as slime-negative (biofilm negative) (5).
3.2.2. Quantitative Method
The microtiter plate (MTP) method was employed to detect bacterial adherence to an abiotic surface. S. aureus strains were grown in tryptic soy broth (TSB) (Merck, Darmstadt, Germany) with 0.25% glucose for 24 hours at 37°C. Then, 200 μL of the bacterial suspension (final concentration ~ 106 CFU/mL) was added to each well of polystyrene MTPs (Greiner, Louis, MO, USA). After 24 hours of incubation at 37°C, the supernatant was discarded, and the wells were washed three times with phosphate-buffered saline (PBS). The adhered cells were stained using 0.3% crystal violet (Merck, Darmstadt, Germany) for 5 - 10 minutes, and then sterile water was used to rinse off the unbound dye. Finally, the stained biofilms were solubilized with an ethanol:acetone (80:20) solution, and absorbance was measured using an ELISA reader (Stat Fax-2100) at 570 nm.
The cut-off OD (ODc) was calculated as three standard deviations (SD) above the mean OD of the negative control: ODc = average OD of negative control + (3 * SD of negative control). The OD value of each strain was defined as the average OD value of the strain reduced by the ODc value (OD = average OD of a strain - ODc). The biofilm formation ability of each strain was categorized as follows: OD570 ≥ 1 = strong biofilm producer, 0.2 ≤ OD570 < 1.0 = moderate biofilm producer, OD570 < 0.2 = weak biofilm producer, OD570 ≤ ODc = biofilm negative.
All experiments were done in triplicate, and the wells without bacterial cells were considered negative controls (5).
3.3. Antimicrobial Susceptibility Testing
The susceptibility of all biofilm-producing strains was tested against 15 antibiotics on Mueller-Hinton agar (MHA) plates (Merck, Germany) using the Kirby-Bauer disk diffusion method according to Clinical Laboratory Standard Institute (CLSI) guidelines. The antibiotic disks used in the study were as follows: Kanamycin (30 µg), nalidixic acid (30 µg), ciprofloxacin (5 µg), levofloxacin (5 µg), ofloxacin (5 µg), norfloxacin (10 µg), streptomycin (10 µg), gentamicin (10 µg), amikacin (30 µg), tobramycin (10 µg), erythromycin (15 µg), azithromycin (15 µg), tetracycline (30 µg), doxycycline (30 µg), and cefoxitin (30 µg). Additionally, the resistance of strains to vancomycin was determined using the broth microdilution assay as recommended by CLSI (17).
3.4. DNA Extraction
The DNA template of all biofilm-positive strains was extracted using a polymerase chain reaction (PCR) template preparation kit (SinaClon, Tehran, Iran) according to the manufacturer's instructions.
3.5. Identification of Staphylococcus aureus Strains
All 119 suspected isolates were identified and confirmed as S. aureus using a PCR assay with species-specific primers for the nucA gene (Appendix 1 in Supplementary File) (18).
3.6. Detection of Biofilm-Related Genes
Biofilm-positive S. aureus strains were screened for the presence of icaA, icaD, cna, fnbpA, and clfA genes using specific primers and PCR conditions mentioned in Appendix 1 in Supplementary File (19).
3.7. Detection of Antibiotic Resistance Genes
Separate PCR reactions were employed to detect aac(6’)-Ie/aph(2”), ant(4’)-Ia, and aph(3’)-IIIa (encoding aminoglycoside-modifying enzymes) (20), as well as gyrA, gyrB, grlA, and grlB (fluoroquinolone resistance) genes, according to the conditions described in Appendix 1 in Supplementary File (12).
3.8. Typing of Bacterial Isolates
The amplification of the mecA gene was carried out using PCR with specific primers for the detection of MRSA strains (18). Moreover, a multiplex-PCR assay was employed to identify various types and subtypes of SCCmec (I, II, III, IVa, IVb, IVc, IVd, and V) among MRSA strains (21). Additionally, the frequency distribution of different agr types (groups I-IV) was determined according to a previously reported protocol for agr typing (19). The sequence of primers and PCR conditions are summarized in Appendix 1 in Supplementary File.
3.9. Biochemical Fingerprinting
Biochemical phenotyping of bacterial strains was carried out using the PhP-CS plates (PhPlate AB, Stockholm, Sweden), which contain four sets of 24 dehydrated reagents. The microplates were incubated at 37°C, and the image of each plate was scanned at 16, 40, and 64-hour intervals with an HP Scanjet 4890 scanner (Hewlett-Packard, Palo Alto, CA, USA). The diversity of the bacterial populations was then calculated using PhPWin software (PhPWin ver. 4.2; PhP Microplate Techniques AB) (22).
3.10. Statistical Analysis
Statistical analyses were performed using GraphPad Prism software (version 8.0). Data for categorical variables were expressed as percentages. Statistical significance was assessed using Fisher’s exact test for categorical variables. Differences were considered statistically significant at P ≤ 0.05.
4. Results
4.1. Identification of Staphylococcus aureus Isolates
All 119 isolates collected from patients with symptomatic UTIs were identified and confirmed as S. aureus strains using specific nucA primers.
4.2. Biofilm Formation
Biofilm formation was evaluated in this study using two different methods: The qualitative CRA and the quantitative MTP methods. On the CRA plates, 12% (N = 14) of S. aureus strains produced slime and black colonies, while 27% (N = 32) were suspected biofilm producers, and 61% (N = 73) were non-biofilm producers. The results of the MTP assay showed that 10 (19%), 68 (57%), and 41 (24%) of S. aureus strains were classified as strong, moderate, and non-biofilm-forming strains, respectively.
4.3. Antibiotic Susceptibility Testing
Results of antibiotic susceptibility testing revealed that the highest antibiotic resistance rate was to nalidixic acid (77%), followed by streptomycin (73%) (Table 1). Additionally, 41%, 36%, 35%, and 32% of strains showed resistance to tobramycin, kanamycin, gentamicin, and amikacin (as aminoglycoside antibiotics), respectively. Furthermore, 36% of strains were resistant to different fluoroquinolone antibiotics tested (ciprofloxacin, ofloxacin, levofloxacin, and norfloxacin). None of the strong biofilm-producing strains were resistant to doxycycline, and 50% of strong and moderate biofilm-forming strains showed resistance to tetracycline. Compared to strong biofilm-forming strains, the rate of resistance to all antibiotics (except for streptomycin, nalidixic acid, and tetracycline) was higher among moderate biofilm-producing isolates. The MICs of different strains to vancomycin were less than 1 µg/mL, indicating susceptibility to vancomycin.
| Antibiotics | Strong Biofilm-Forming Strains | Moderate Biofilm-Forming Strains | Total | P-Value |
|---|---|---|---|---|
| FOX | 3 (30) | 31 (46) | 34 (44) | 0.0286 |
| E | 3 (30) | 31 (46) | 34 (44) | 0.0286 |
| AZM | 4 (40) | 32 (47) | 36 (46) | 0.3922 |
| D | 0 | 4 (6) | 4 (5) | 0.0289 |
| TE | 5 (50) | 34 (50) | 39 (50) | 1.0000 |
| NA | 8 (80) | 52 (76) | 60 (77) | 0.6089 |
| CIP | 2 (20) | 26 (38) | 28 (36) | 0.0078 |
| LEV | 2 (20) | 26 (38) | 28 (36) | 0.0078 |
| OFX | 2 (20) | 26 (38) | 28 (36) | 0.0078 |
| NOR | 2 (20) | 26 (38) | 28 (36) | 0.0078 |
| S | 8 (80) | 49 (72) | 57 (73) | 0.2463 |
| GM | 2 (20) | 25 (37) | 27 (35) | 0.0118 |
| TOB | 3 (30) | 29 (43) | 32 (41) | 0.0776 |
| AN | 2 (20) | 23 (34) | 25 (32) | 0.0378 |
| K | 2 (20) | 26 (38) | 28 (36) | 0.0078 |
| VAN | 0 | 0 | 0 | - |
Abbreviations: AN, amikacin; AZM, azithromycin; CIP, ciprofloxacin; D, doxycycline; E, erythromycin; FOX, cefoxitin; GM, gentamicin; K, kanamycin; LEV, levofloxacin; NA, nalidixic acid; NOR, norfloxacin; OFX, ofloxacin; S, streptomycin; TE, tetracycline; TOB, tobramycin; VAN, vancomycin.
a Values are expressed as No. (%) unless otherwise indicated.
Strong and moderate biofilm-producing strains were categorized into 8 and 32 antibiotypes, respectively (Table 2). The most frequent resistance type was antibiotype 35 (resistance to 14 different antibiotics), occurring in 24% (N = 19) of isolates. Additionally, 6% (N = 4) of the moderate biofilm producers were susceptible to all studied antibiotics (Table 2). In total, 40% of strong biofilm-producer strains and 36% of moderate biofilm-producer strains were resistant to at least one antibiotic in three or more classes of antibiotics, categorizing them as multidrug-resistant (MDR) strains. Moreover, 34 strains (44%) showed resistance to cefoxitin and were positive for the mecA gene, identifying them as MRSA.
| Antibiotics | Pattern | Strong Biofilm Producing | Moderate Biofilm Producing | Total |
|---|---|---|---|---|
| No antibiotic | ||||
| 0 | 1 | - | 4 (6) | 4 (5) |
| One antibiotic | ||||
| NA | 2 | - | 5 (7) | 5 (6) |
| S | 3 | 1 (10) | 5 (7) | 6 (8) |
| AZM | 4 | - | 1 (1.45) | 1 (1.2) |
| TE | 5 | 1 (10) | - | 1 (1.2) |
| Two antibiotics | ||||
| NA, TE | 6 | 1 (10) | - | 1 (1.2) |
| NA, S | 7 | 2 (20) | 9 (13) | 11 (14) |
| NA, FOX | 8 | - | 1 (1.45) | 1 (1.2) |
| NA, TOB | 9 | - | 1 (1.45) | 1 (1.2) |
| S, FOX | 10 | - | 1 (1.45) | 1 (1.2) |
| TE, FOX | 11 | - | 1 (1.45) | 1 (1.2) |
| Three antibiotics | ||||
| NA, S, TE | 12 | - | 2 (3) | 2 (3) |
| NA, S, AZM | 13 | 1 (10) | - | 1 (1.2) |
| NA, AZM, E | 14 | - | 1 (1.45) | 1 (1.2) |
| NA, S, TOB | 15 | 1 (10) | 1 (1.45) | 2 (3) |
| NA, S, FOX | 16 | - | 1 (1.45) | 1 (1.2) |
| S, AN, TE | 17 | - | 1 (1.45) | 1 (1.2) |
| S, AZM, E | 18 | - | 1 (1.45) | 1 (1.2) |
| TE, D, FOX | 19 | - | 1 (1.45) | 1 (1.2) |
| Four antibiotics | ||||
| NA, AZM, TE, E | 20 | - | 1 (1.45) | 1 (1.2) |
| NA, S, TOB, TE | 21 | - | 1 (1.45) | 1 (1.2) |
| Five antibiotics | ||||
| NA, S, K, TOB, TE | 22 | - | 1 (1.45) | 1 (1.2) |
| NA, S, AZM, E, FOX | 23 | - | 1 (1.45) | 1 (1.2) |
| Six antibiotics | ||||
| NA, S, AZM, E, TE, FOX | 24 | 1 (10) | - | 1 (1.2) |
| Seven antibiotics | ||||
| NA, GM, K, TOB, AZM, E, TE | 25 | - | 1 (1.4) | 1 (1.2) |
| Nine antibiotics | ||||
| NA, S, GM, K, TOB, AZM, E, TE, FOX | 26 | - | 1 (1.45) | 1 (1.2) |
| Ten antibiotics | ||||
| NA, S, LEV, CIP, OFX, NOR, AZM, E, TE, FOX | 27 | - | 1 (1.45) | 1 (1.2) |
| NA, S, LEV, CIP, OFX, NOR, AZM, E, TE, D | 28 | 1 (1.45) | 1 (1.2) | |
| Eleven antibiotics | ||||
| NA, S, LEV, CIP, OFX, NOR, AZM, E, TE, D, FOX | 29 | - | 1 (1.45) | 1 (1.2) |
| Twelve antibiotics | ||||
| NA, S, LEV, CIP, OFX, NOR, AZM, E, GM, K, TOB, FOX | 30 | - | 1 (1.45) | 1 (1.2) |
| NA, S, LEV, CIP, OFX, NOR, GM, AN, TE, K, TOB, FOX | 31 | - | 1 (1.45) | 1 (1.2) |
| NA, LEV, CIP, OFX, NOR, GM, K, AN, TOB, AZM, E, FOX | 32 | - | 1 (1.45) | 1 (1.2) |
| LEV, CIP, OFX, NOR, GM, K, AN, TOB, AZM, E, TE, FOX | 33 | - | 1 (1.45) | 1 (1.2) |
| Thirteen antibiotics | ||||
| NA, S, LEV, CIP, OFX, NOR, GM, AN, AZM, K, E, TE, TOB | 34 | - | 1 (1.45) | 1 (1.2) |
| Fourteen antibiotics | ||||
| NA, S, LEV, CIP, OFX, NOR, GM, K, AN, TOB, AZM, R, TE, FOX | 35 | 2 (20) | 17 (25) | 19 (24) |
| Fifteen antibiotics | ||||
| NA, S, LEV, CIP, OFX, NOR, GM, K, AN, TOB, AZM, R, TE, D, FOX | 36 | - | 1 (1.45) | 1 (1.2) |
Abbreviations: AN, amikacin; AZM, azithromycin; CIP, ciprofloxacin; D, doxycycline; E, erythromycin; FOX, cefoxitin; GM, gentamicin; K, kanamycin; LEV, levofloxacin; NA, nalidixic acid; NOR, norfloxacin; OFX, ofloxacin; S, streptomycin; TE, tetracycline; TOB, tobramycin; VAN, vancomycin.
a Values are expressed as No. (%).
4.4. Biofilm Related Genes
Results of PCR using specific primers showed that the icaA and icaD genes were present in 78% and 82% of strains, respectively. Additionally, the prevalence of the clfA, fnbA, and cna genes was 68%, 59%, and 42% of strains, respectively (Figure 1). Among the 10 strong biofilm-producing strains, 80% harbored both icaA and icaD genes. These frequencies among moderate biofilm formers were 78% and 82%, respectively. Moreover, the frequency of clfA and cna among moderate biofilm-producing strains was significantly higher (P < 0.0001) than among strong biofilm-forming strains.
4.5. Antibiotic Resistance Genes
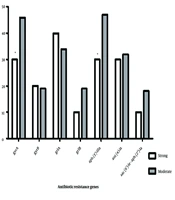
Among aminoglycoside-modifying enzymes, the aph(3′)-IIIa gene was the dominant gene, detected in 45% of strains, followed by the ant(4)-Ia (32%) and aac(6')-Ie-aph(2'')-Ia (17%) genes (Figure 2). The aph(3′)-IIIa gene was the most frequent among strong (30%) and moderate (47%) biofilm-producing strains, while the aac(6')-Ie-aph(2'')-Ia variant was the least detected AME-encoding gene type.
Among quinolone resistance genes, gyrA and grlA were the most prevalent among all biofilm-producing strains, detected in 44% and 35% of strains, respectively (Figure 2). The frequency of grlA (40%) and gyrB (20%) among strong biofilm-producing strains was higher than among moderate biofilm-forming ones. Conversely, gyrA and grlB were the most frequent genes among moderate biofilm formers.
4.6. SCCmec and agr Typing
Among all 34 MRSA strains, five different SCCmec types were detected: Types I, III, IVa, IVc, and V. SCCmec type III was present in 64% (N = 22) of the strains (Table 3). Additionally, 24% (N = 8) and 76% (N = 26) of the strains were classified as community-acquired MRSA (CA-MRSA) and hospital-acquired MRSA (HA-MRSA), respectively. Out of the three strong biofilm-forming MRSA strains, two (67%) harbored SCCmec type III and one (33%) harbored SCCmec type IVc. SCCmec type III (65%, N = 20) was the most prevalent among moderate biofilm-producing strains, followed by types V (13%, N = 4), I (13%, N = 4), IVa (6%, N = 2), and IVc (3%, N = 1).
| Variables and Types | Strong Biofilm Producing | Moderate Biofilm Producing | Total | P-Value |
|---|---|---|---|---|
| SCCmec | ||||
| Type I | - | 4 (13) | 4 (12) | 0.0002 |
| Type III | 2 (67) | 20 (65) | 22 (64) | 0.8814 |
| Type IVa | - | 2 (6) | 2 (6) | 0.0289 |
| Type IVc | 1 (33) | 1 (3) | 2 (6) | < 0.0001 |
| Type V | - | 4 (13) | 4 (12) | 0.0002 |
| agr | ||||
| Type I | 3 (30) | 32 (47) | 35 (45) | 0.0198 |
| Type II | 1 (10) | 2 (3) | 3 (4) | 0.0818 |
| Type III | 3 (30) | 5 (7) | 8 (10) | < 0.0001 |
| Type IV | - | 1 (1) | 1 (1) | 1.0000 |
| Non typeable | 3 (30) | 28 (42) | 31 (40) | 0.1048 |
a Values are expressed as No. (%) unless otherwise indicated.
All four agr types were successfully identified in 47 strains (60%), with agr type I being the dominant type among all biofilm-producing strains (Table 3). Furthermore, 31 (40%) strains were negative for all agr types and classified as non-typeable. agr type I (47%) was the predominant type among moderate biofilm-forming strains, followed by agr types III (7%), II (3%), and IV (1%). Molecular detection of 10 strong biofilm-forming S. aureus isolates indicated that agr types I (30%) and III (30%) were the most prevalent, followed by agr type II (10%).
4.7. Clonality of Bacterial Strains
Phene plate typing of bacterial strains revealed the presence of 22 PhP types, consisting of 8 common types (CTs) and 14 single types (STs). CT2 contained the highest number of bacterial strains (N = 31, 40%), followed by CT3 (N = 13, 17%) and CT4 (N = 8, 10%) (Table 4 and Appendix 2 in Supplementary File). While the prevalence of strong biofilm-producing strains was limited to CTs 1, 2, 3, and 5, CTs 4, 6, 7, and 8 were only common among moderate biofilm-forming S. aureus strains. Among the 14 STs detected, STs 2, 4, 5, and 7 belonged to strong biofilm producers, and the remaining STs were limited to moderate biofilm formers.
| No. | Phene plate Type | Antibiotype | Biofilm Formation | agr Type | mecA | SCCmec Type |
|---|---|---|---|---|---|---|
| 1 | CT1 | 5 | Strong | - | - | - |
| 2 | CT1 | 4 | Moderate | 4 | - | - |
| 3 | CT1 | 1 | Moderate | - | - | - |
| 4 | CT1 | 35 | Moderate | 1 | + | III |
| 5 | ST1 | 14 | Moderate | - | - | - |
| 6 | CT2 | 33 | Moderate | - | + | V |
| 7 | CT3 | 7 | Strong | 1 | - | - |
| 8 | ST2 | 13 | Strong | 3 | - | - |
| 9 | CT2 | 7 | Moderate | - | - | - |
| 10 | CT3 | 7 | Moderate | 1 | - | - |
| 11 | CT2 | 3 | Moderate | 1 | - | - |
| 12 | CT2 | 29 | Moderate | 1 | + | III |
| 13 | CT3 | 35 | Moderate | 1 | + | III |
| 14 | CT2 | 27 | Moderate | 1 | + | III |
| 15 | CT2 | 2 | Moderate | - | - | - |
| 16 | CT2 | 7 | Moderate | 1 | - | - |
| 17 | CT3 | 2 | Moderate | - | - | - |
| 18 | CT3 | 28 | Moderate | 1 | - | - |
| 19 | CT3 | 30 | Moderate | - | + | V |
| 20 | CT3 | 31 | Moderate | 1 | + | I |
| 21 | ST3 | 1 | Moderate | - | - | - |
| 22 | CT2 | 35 | Moderate | 1 | + | III |
| 23 | CT3 | 35 | Moderate | 1 | + | III |
| 24 | CT2 | 35 | Moderate | - | + | III |
| 25 | ST4 | 24 | Strong | 3 | + | IVc |
| 26 | ST5 | 3 | Strong | 3 | - | - |
| 27 | ST6 | 2 | Moderate | 1 | - | - |
| 28 | CT2 | 11 | Moderate | 1 | + | V |
| 29 | CT3 | 3 | Moderate | 3 | - | - |
| 30 | CT2 | 6 | Strong | 1 | - | - |
| 31 | ST7 | 7 | Strong | 2 | - | - |
| 32 | CT2 | 34 | Moderate | - | - | _ |
| 33 | CT4 | 35 | Moderate | 1 | + | III |
| 34 | CT2 | 25 | Moderate | 3 | - | - |
| 35 | CT2 | 2 | Moderate | 3 | - | - |
| 36 | CT2 | 1 | Moderate | 1 | - | - |
| 37 | CT2 | 20 | Moderate | 1 | - | - |
| 38 | CT4 | 19 | Moderate | 3 | + | I |
| 39 | CT3 | 7 | Moderate | - | - | - |
| 40 | CT5 | 35 | Strong | - | + | III |
| 41 | CT2 | 35 | Moderate | 1 | + | III |
| 42 | CT3 | 35 | Strong | 1 | + | III |
| 43 | CT6 | 35 | Moderate | 1 | + | III |
| 44 | CT6 | 2 | Moderate | - | - | - |
| 45 | CT3 | 1 | Moderate | - | - | - |
| 46 | CT2 | 8 | Moderate | 1 | + | IVa |
| 47 | CT2 | 3 | Moderate | - | - | - |
| 48 | CT3 | 3 | Moderate | 1 | - | - |
| 49 | CT2 | 7 | Moderate | 1 | - | - |
| 50 | CT7 | 3 | Moderate | - | - | - |
| 51 | CT8 | 7 | Moderate | 1 | - | - |
| 52 | CT2 | 35 | Moderate | 1 | + | III |
| 53 | CT2 | 35 | Moderate | 1 | + | IVc |
| 54 | CT4 | 35 | Moderate | 1 | + | III |
| 55 | CT8 | 16 | Moderate | 1 | + | I |
| 56 | ST8 | 10 | Moderate | - | + | III |
| 57 | ST9 | 22 | Moderate | - | - | - |
| 58 | CT2 | 35 | Moderate | 1 | + | III |
| 59 | ST10 | 35 | Moderate | 1 | + | III |
| 60 | CT2 | 7 | Moderate | 3 | - | - |
| 61 | CT2 | 7 | Moderate | 1 | - | - |
| 62 | CT2 | 35 | Moderate | - | + | III |
| 63 | ST11 | 35 | Moderate | 1 | + | III |
| 64 | CT2 | 35 | Moderate | 1 | + | III |
| 65 | CT4 | 15 | Moderate | 2 | - | - |
| 66 | CT7 | 9 | Moderate | 2 | - | - |
| 67 | CT4 | 18 | Moderate | - | - | - |
| 68 | CT2 | 12 | Moderate | - | - | - |
| 69 | CT2 | 17 | Moderate | - | - | - |
| 70 | CT4 | 21 | Moderate | - | - | - |
| 71 | CT2 | 15 | Strong | - | - | - |
| 72 | CT2 | 35 | Moderate | 1 | + | III |
| 73 | CT4 | 32 | Moderate | - | + | IVa |
| 74 | ST12 | 36 | Moderate | - | + | III |
| 75 | CT5 | 23 | Moderate | - | + | V |
| 76 | ST13 | 7 | Moderate | - | - | - |
| 77 | ST14 | 12 | Moderate | - | - | - |
| 78 | CT4 | 26 | Moderate | - | + | I |
5. Discussion
Staphylococcus aureus is considered a significant threat to patients with UTIs due to its potential to invade renal tissues by adhering to the urothelium as a result of biofilm formation (23). Therefore, reliable, convenient, accessible, and inexpensive methods are needed to identify biofilm-forming strains (24). In this study, biofilm-producing strains were detected using a combination of the Congo-red agar plate test as a primary screening method and the MTP assay as the gold standard test for quantitative evaluation of biofilm formation. Our results showed that 76% of isolates were biofilm producers, which is consistent with the study carried out by Yousefi et al. in Iran (1).
To understand the molecular mechanism of biofilm formation in S. aureus, we aimed to detect five key genes involved in this process. According to our results, icaD and icaA were the most prevalent genes. Several reports have indicated that the biofilm formation capacity in S. aureus causing UTIs is associated with the presence of icaABCD genes (23, 25), mentioning these genes as essential factors for intercellular adhesion and multilayer bacterial biofilm production (26). Moreover, in a study in China, the icaABCD genes were detected in all S. aureus strains (27). The biofilm-forming capacity of some isolates in the absence of the icaAD genes indicated the importance of further genetic investigations into ica-independent biofilm formation mechanisms. In the ica-independent pathway, different proteins including FnbpA, ClfA/B, and collagen-binding adhesin (CNA) may be involved in biofilm formation, as detected in this study.
In this study, 44% of biofilm-producing strains isolated from patients with UTIs were categorized as MRSA. In similar surveys conducted in Iraq and Australia, the prevalence of MRSA strains among urine specimens was 7.7% and 4.06%, respectively (28, 29). Different frequencies of MRSA strains have been reported from various sources in Iran, ranging from 19% to 70%, indicating the dissemination and persistence of different MRSA clone types in the country (30, 31). The increase in resistance to cefoxitin in the present study could be attributed to bacterial evolution, excessive antibiotic consumption, antibiotic abuse, self-treatment, and the transmission of resistant strains from the environment, animals, foods, and between individuals.
Our results revealed that the highest antibiotic resistance rates were against nalidixic acid and streptomycin. This finding contrasts with previous findings by Khaleel et al. in Iraq, who reported that S. aureus strains from UTIs were highly resistant to penicillin (100%), ceftaroline (100%), and gentamicin (87.2%) (28). Another study conducted in Iran showed high resistance of S. aureus to tetracycline, ciprofloxacin, and erythromycin (1). Despite the high resistance rates to different antibiotics tested, resistance to doxycycline was relatively low and uncommon among biofilm-producing isolates in this study. Therefore, this antibiotic can be used as an appropriate treatment for UTIs caused by biofilm-forming S. aureus strains.
Previous studies conducted in Nigeria showed that ciprofloxacin was the most effective antibiotic against S. aureus strains isolated from UTIs (32). Alshomrani et al. reported that 91% of S. aureus isolated from urine in Saudi Arabia were susceptible to trimethoprim-sulfamethoxazole (33). Treatment of UTIs is usually started empirically before the preparation of laboratory results of urine culture. However, the prevalent causes of the infection and their antibiotic susceptibility patterns show wide geographic variation. Therefore, we recommend that empirical antibiotic selection be based on knowledge of local patterns of urinary pathogens and their antibiotic sensitivities.
The most common resistance mechanism against aminoglycosides in staphylococci is drug inactivation by cellular aminoglycoside-modifying enzymes, including acetyltransferase (AAC), adenylyltransferase, and phosphotransferase (APH) (12). Unlike other studies (14, 34, 35), the aac(6')-Ie-aph(2'')-Ia gene had the lowest prevalence among aminoglycoside-resistant (AMR) strains in our study, indicating the emergence of different clone types in the studied hospital that are distinct from previous clones circulating in Iran and other countries.
Mechanisms of fluoroquinolone resistance in S. aureus are mostly related to mutations in drug targets, including DNA topoisomerase IV (encoded by grlA and grlB) and DNA gyrase (encoded by gyrA and gyrB). Our results showed that gyrA and grlA were the most prevalent genes among quinolone-resistant strains. Compared to previous reports from Iran, the frequency of these genes was higher among biofilm-producing strains (34, 36). This genetic diversity may be due to the presence of multiple sources of resistant bacteria or the possible exchange of genes among different strains through the transfer of mobile genetic elements (34).
The presence of an antibiotic resistance phenotype is mostly associated with the presence of corresponding antibiotic resistance genes (36). Our results indicated that the incidence of a phenotypic profile of quinolone resistance (36%) is almost equal to the incidence of a genotypic profile (gyrA; 44% and grlA 35%). Thus, the expression of such genes under suitable conditions may potentially lead to therapeutic failure.
Few studies describe the prevalence and dissemination of various clonal groups of biofilm-forming S. aureus strains among patients with UTIs in Iran. In this study, the majority of bacterial strains harbored SCCmec type III. Previous reports have supported our findings, indicating that SCCmec type III is the predominant type in Iran (5, 22, 37), suggesting the hospital origin of such strains. However, other studies have shown that biofilm-producing S.aureus strains were mostly SCCmec type IV (11, 38). In the present research, agr type I was the dominant type among all biofilm-producing strains. These results contrast with previous studies indicating that isolates with agr types II and III produced large amounts of biofilm due to a deficiency in icaR transcription, which acts as a repressor of the icaADBC operon (5, 39). Although our findings align with other research showing that isolates belonging to agrI had a great ability to form strong biofilms (40), some studies have indicated the absence of an association between specific agr genotypes and enhanced ability to form biofilm (41).
Compared to genotyping methods such as pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST), the PhP system is specifically designed for each bacterial species to yield the highest discrimination among the strains of that species (14). Our results showed the presence of diverse PhP types (22 types) among biofilm-producing strains, consisting of 8 CTs and 14 STs, indicating high diversity in the kinetics of metabolism of strains. CT2 was the predominant type, and its dissemination is consistent with previous studies in Iran (5, 14). The presence of different antibiotypes and biofilm formation abilities among the strains with the identical PhP type indicated no relatedness between their clonal dissemination, biofilm production, and antibiotic resistance profiles.
Despite the valuable insights provided by this study, several limitations may impact the interpretation of our results. First, there were relatively long time intervals between sample collection, data analysis, and reporting of results. Second, the study focused on a particular geographical zone for the selection of patients. Therefore, these results should be interpreted with caution, as they may not fully represent the prevalence, diversity, and antimicrobial susceptibility profiles of S. aureus strains among patients with UTIs in this country at the present time.
5.1. Conclusions
Our results revealed the presence of various biofilm production capacities, antimicrobial resistance profiles, and clonal lineages in S. aureus isolated from patients with UTIs. Considering the significant role of S. aureus in human medicine, the carriage of biofilm-related and antibiotic resistance genes provides evolutionary benefits to this microorganism, potentially leading to the selection of more resistant strains in the future. The biofilm-forming capacity of both MRSA and MSSA strains indicates a high ability of these strains to persist in hospital environments, increasing the risk of disease development in hospitalized patients. Therefore, continuous tracking of epidemic strains and adopting novel approaches for inhibiting biofilm formation are recommended to control the dissemination of bacteria among inpatients effectively. Additionally, our results suggest that doxycycline can be prescribed in the empirical therapy of UTIs caused by biofilm-forming S. aureus strains. We recommend future research to determine the accurate role of other virulence factors and their correlation to the pathogenic potential of S. aureus and the development of drug resistance to develop better therapeutic measures against S. aureus infections.