1. Background
COVID-19, caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), is a disease characterized by a high level of transmissibility. It has affected 226 countries and continents, resulting in a global death toll exceeding 6.2 million individuals. Despite the collective efforts of scientific institutions worldwide to identify potential therapeutic interventions, the Food and Drug Administration (FDA) has not yet approved any specific pharmaceutical agent for the treatment of COVID-19. The emergence of SARS-CoV-2 variants of concern significantly reduces the likelihood of existing therapeutics effectively addressing and detecting the onset of the disease (1). The etiology of SARS-CoV-2 infection and the dynamics of virus-host interactions in the context of COVID-19 remain incomplete and require further elucidation. Therefore, investigating the biochemical and molecular causes of disease variation is crucial. Omics methods play a pivotal role in providing essential data for public health policy decision-making and developing novel biomarkers to predict disease severity in COVID-19 patients (2).
MicroRNAs (miRNAs) are a class of small non-coding RNA molecules that regulate gene expression. They perform this regulatory function by binding to the 3´ untranslated regions of target RNAs. Innate and adaptive immunity, as well as the fine-tuning of immune responses, rely heavily on this post-transcriptional control. Numerous miRNAs regulate inflammation-related mediators, making them indispensable in certain inflammatory diseases (3). Additionally, substantial evidence underscores the critical role of miRNAs in regulating inflammatory mediators, highlighting their importance in the pathogenesis of specific inflammatory disorders. MicroRNAs have been shown to play essential roles in the pathogenesis and treatment of various viral infections, including respiratory syncytial virus (RSV), Middle East respiratory syndrome coronavirus (MERS-CoV), severe acute respiratory syndrome (SARS), and SARS-CoV-2 (4-7).
Many microRNAs from the host have been shown to target the SARS-CoV-2 genome, suppressing viral production and reducing pathogenicity. Specific mutations in the 3´UTR region of the viral genome enable the virus to evade the host immune system, which can influence the outcome of SARS-CoV-2 infection through its interaction with the cellular miRNome. Determining the miRNA profile associated with various severity outcomes, particularly at the onset of the illness, may provide valuable insights for improving the diagnosis and treatment of COVID-19 (6, 8).
MicroRNAs may play a crucial role in mitigating the inflammatory response induced by SARS-CoV-2 (9). They currently have a significant regulatory impact on cytokines, chemokines, and growth factors (10-12). This regulation is directly linked to the occurrence of cytokine release syndrome (CRS) in individuals affected by COVID-19. Analyzing these miRNAs could lead to the identification of promising therapeutic targets to combat CRS and uncover potential miRNA biomarkers for predicting survival outcomes at the onset of the disease (13).
2. Objectives
According to a recent study employing software and bioinformatics analyses, miRs 1307-3p and 3613-5p have been proposed to impede viral replication by targeting the 3´UTR region of the viral genome (14). Given the widespread dissemination of the COVID-19 virus, this research examined the expression levels of two specific human miRs, which hold potential as biomarkers for detecting and identifying COVID-19 infection. Consequently, the findings of the present study provide a foundation for future investigations into the antiviral properties of these miRs and their potential applications as antiviral therapies.
3. Methods
3.1. Clinical Samples
Twenty-six whole blood samples were obtained from hospitalized patients with confirmed COVID-19 infection, as determined by real-time PCR testing of their swab samples between October and December 2020. The mean age of the patients was 54.4 ± 14.3 years, with 15 men (57.7%) and 11 women (42.3%). Except for one patient, who was 19 years old, all others were older than 35, with the oldest being 76 years old. An equal number of healthy individuals were selected as the control group, matched accordingly. The collected samples were subjected to PBMC separation using a Ficoll solution.
The selection of individuals for the study groups was based on their COVID-19 test results. Individuals hospitalized with clinical symptoms of COVID-19, such as fever, shortness of breath, cough, myalgia, nausea, chest pain, and positive RT-PCR test results from swab samples, were included in the patient group. Conversely, those with negative RT-PCR test results and no respiratory symptoms were included in the control group.
3.2. Sample Preparation
Five-milliliter whole blood samples were obtained from human subjects with utmost care and placed in individual sterile test tubes with screw caps (Iran) containing EDTA as an anticoagulant. To dilute the blood, 5 mL of PBS was added, and the mixture was transferred to a Falcon tube containing 2.5 mL of Ficoll solution for PBMC isolation. The tube was centrifuged at 2000 rpm for 20 minutes. The buffy coat layer was carefully isolated, added to a microtube containing lysis buffer, and stored at -80°C, ensuring the ethical handling of the samples.
3.3. The Process of Isolating MicroRNA
The RNX solution (SinaClon, Iran) was used to extract total RNA via the isopropanol-chloroform precipitation method. RNA yield and purity were assessed using a NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE) by measuring absorbance at 260 and 280 nm. Only samples with a 260/280 ratio of 1.7 or higher were included in the analysis. The isolated total RNA samples were stored at -80°C until further use.
3.4. Quantitative Reverse Transcription-Polymerase Chain Reaction
Complementary DNA (cDNA) for each miRNA was synthesized using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Grand Island, NY) and miRNA-specific stem-loop primers, following the manufacturer's instructions. The stem-loop structure facilitates the reverse transcription of the mature miRNA into cDNA at the 5'-end of the primer, designed to prevent primers from annealing to other mature miRNA variants or miRNA precursors. All cDNA samples were stored at -20°C until reverse transcription-polymerase chain reaction (RT-qPCR) analysis.
Reverse transcription-polymerase chain reaction was performed on an ABI7500 system using the TaqMan miRNA Assay (Applied Biosystems) to quantify mature miRNAs. The PCR reaction mixture consisted of 1 µL of gene-specific TaqMan miRNA real-time PCR Assay primer (Applied Biosystems), 20 µL of TaqMan 2x Universal PCR Master Mix, and 9 µL of miRNA-specific cDNA. The thermal cycling conditions included activation of Taq polymerase at 95°C for 10 minutes, followed by 40 cycles of PCR amplification at 95°C for 20 seconds and annealing/elongation at 60°C for 1 minute.
MicroRNA expression levels were normalized using Snord as a reference. MicroRNA expressions were compared using the 2-ΔΔCt method. Table 1 provides the sequences of the primers used for RT-qPCR.
| Gene Name | Forward Sequence (5'-3') |
|---|---|
| 1307-3p loop | GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACCACGAC |
| Universal Reverse | GTGCAGGGTCCGAGGT |
| 1307-3p-F | TTGTACTCGGCGTGGCGT |
| 3613-5p loop | GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACGAACAA |
| 3613-5p-F | GGGGGGTGTTGTACTTTTTTTT |
| Snord 48 | GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACGGTCAG |
| Snord 48-F | GTAGTGATGATGACCCCAG |
The Primer Sequences Employed for Quantitative Polymerase Chain Reaction
3.5. Statistical Analysis
The statistical significance of the data was evaluated using a t-test performed with GraphPad Prism 8 software (GraphPad Software, Inc., USA). A P-value of less than 0.05 was considered statistically significant. The data are presented as the mean ± standard deviation (SD).
4. Results
4.1. Reverse Transcription-Polymerase Chain Reaction Study Identifies MicroRNA as COVID-19 Markers
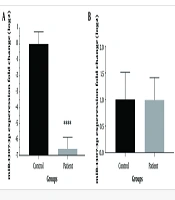
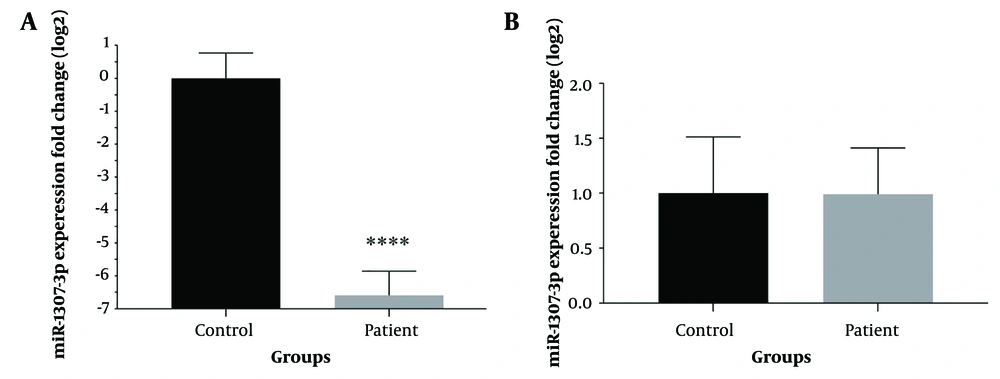
To identify significant differences in miRNA expression between individuals with COVID-19 and healthy controls, we analyzed the expression levels of two specific miRNAs selected based on previous research findings (14). The results from the RT-qPCR analysis revealed a significant reduction in the expression level of miR-1307-3p (P < 0.0001) in the patient group compared to the control group. In contrast, no significant differences were observed in the levels of miR-3613-5p between the patient and control groups (Figure 1).
5. Discussion
In most cases, widespread pandemics result from the emergence of a previously unknown virus. SARS-CoV-2 triggered a deadly pandemic in China in December 2019. Given the severity of the situation, a rapid and comprehensive study of SARS-CoV-2 was essential (14). While the origin of the coronavirus remains a topic of debate, it is well established that certain viruses, such as COVID-19, can be transmitted freely between humans and animals.
When a virus or other infectious agent enters the body, it can alter various cellular processes, including miRNA expression. Human infectious diseases, particularly zoonotic ones, present substantial research gaps, and the role of miRNAs in regulating host-pathogen interactions remains largely unexplored. Therefore, further in-depth investigations are required to understand their characteristics and interactions with viruses, particularly SARS-CoV-2 (15).
MicroRNAs play a critical role in modulating immune system function and the progression of SARS-CoV-2 infection. These molecules can disrupt SARS-CoV-2 infection through several mechanisms. First, cellular miRNAs can interfere with the virus's entry phase by modulating their expression. Second, miRNAs derived from the viral genome can compete with cellular RNAs, thereby reducing their expression. Third, cellular miRNAs can influence the replication of the viral genome or inhibit virus-encoded RNAs that suppress the expression of specific cellular proteins (16).
In response to viral infection, host cells begin synthesizing miRs, activating their antiviral roles through two distinct mechanisms. The expression of genes essential for viral replication can be modulated in one of two ways: Either by targeting the virus directly or by regulating immune system genes. These processes disrupt the virus's ability to replicate its genome or produce proteins (17). Recent studies have shown that viral RNA can mimic miRNAs or ribosomal RNA (rRNA) and bind to host cell mRNA (18, 19).
An interesting study revealed that sequences highly similar to cellular miR-1307-3p and other miRNAs are present in the genomes of various coronavirus strains, including SARS-CoV-2 (20). Research using laboratory models has confirmed that sequences resembling miR-1307 within sections of the SARS-CoV-2 genome can bind to this miRNA, benefiting the viral replication cycle (21). Based on Chen and Zhong’s investigation, this research analyzed the expression levels of two microRNAs, miR-1307-3p and miR-3613-5p, through bioinformatics analysis, which indicated their potential influence on the SARS-CoV-2 genome (14).
Despite a significant decrease in miR-1307-3p expression, no statistically significant change in miR-3613-5p expression was observed between the control and patient groups in this study. Previous research has shown that miR-1307-3p is among the RNAs associated with lung tissue (22). The expression of this miRNA is linked to various intracellular pathways, including the TGF-β pathway and inflammatory responses. Given the role of inflammation and inflammatory processes in the progression and severity of COVID-19, a significant connection can be established between the expression of this miRNA and viral infection (23).
Another study demonstrated that miR-1307 expression could inhibit the clathrin-mediated endocytosis process, a mechanism that counters the viral replication cycle (24). Zheng et al. conducted a comprehensive analysis of various miRNA expressions in dogs infected with H5N1, further supporting the importance of miRNAs in host-pathogen interactions (25). Dogs infected with H5N1 influenza exhibited reduced expression of miR-1307 and miR-1346, which specifically target SOCS proteins. SOCS family members play a critical role in regulating the JAK-STAT signaling pathway, and their involvement in various diseases has been the focus of recent research. Virus-induced SOCS can block the interferon signaling pathway (25). miR-1307-3p may have a significant role in promoting the production of various interleukins and their receptors in individuals with severe COVID-19 (26).
Analysis of clinical samples also indicates that the expression level of this miRNA in lung tissue increases during SARS-CoV-2 infection (20). In a recent study, Arisan et al. made an intriguing discovery. They observed that the expression of miR-1307-3p in Vero cells increased following infection with SARS-CoV-2. Furthermore, when anti-miR-1307 was introduced to suppress its levels, an increase in the cell survival rate was noted. This finding suggests that inhibiting miR-1307-3p may promote viral replication. Consequently, reducing the expression of this miRNA could mitigate the adverse effects of viral infection on the cell (26).
Similarly, Chen et al. demonstrated an increase in miR-1307 expression after infection in Calu-3 cells (27). These conflicting results regarding the expression of miR-1307 suggest that factors such as tissue or cell conditions and the experimental context (in vivo or in vitro) can significantly influence miRNA expression outcomes.
Two of the mentioned studies reported the expression of miR-1307-3p in Vero and Calu-3 cell lines. However, the present study evaluated the expression of miR-1307-3p in PBMC cells from hospitalized COVID-19 patients. According to the study by Arisan et al., lowering the expression of miR-1307 can increase cell survival rates, which is a process that benefits the virus. Similarly, Zheng et al.'s study showed that reducing the expression of this miRNA can enhance the performance of SOCS, leading to the suppression of the interferon signaling pathway—a process that also supports viral replication (25, 26).
As previously mentioned, regions within the viral genome resembling miR-1307 can neutralize and reduce the expression of cellular miR-1307-3p. However, further studies are needed on patients with COVID-19 exhibiting different severity levels to deepen our understanding of these mechanisms. The small sample size is a notable limitation of this study. Due to the emergency conditions during the COVID-19 pandemic, access to hospitalized patients in ICU wards was restricted.