1. Background
Urinary tract infections (UTIs) are among the most prevalent infectious diseases in children and are a leading cause of antibiotic use and pediatric hospitalization (1, 2). Urinary tract infections also represent a significant challenge for pediatricians, healthcare providers, and the management of children in emergency departments (1, 3, 4). Bacteria are the primary cause of UTIs, with fungi and some viruses being rare contributors. Uropathogenic Escherichia coli is the most frequent cause of UTIs, followed by Staphylococcus species, Klebsiella pneumoniae, Proteus mirabilis, and Enterococcus faecalis (4).
Escherichia coli strains that cause UTIs have defense mechanisms, including a glycosylated polysaccharide capsule that protects against phagocytosis and the immune complement system (2). Escherichia coli is one of the most significant causative agents of UTIs, cholecystitis, bacteremia, meningitis, and traveler's diarrhea (5). More than 40% of the bacteria responsible for UTIs are resistant to some of the antibiotics used, leading to patient relapse, treatment failure, progression to more severe diseases (such as bacteremia), which requires hospitalization, and, ultimately, death (3).
Gram-negative bacteremia remains a leading cause of mortality and morbidity, with its incidence rising globally (6). The presence of resistant genes on extrachromosomal DNA, primarily found in the Enterobacteriaceae family, is mainly responsible for the occurrence of antibiotic resistance in gram-negative bacteria that cause infections in hospitals and the community (7).
The second most common cause of death among young children is diarrhea, often resulting from pathogens transmitted from the mouth to the feces (8). Escherichia coli (enteropathogenic, enteroinvasive, and enteroaggregative strains) is one of the causes of diarrhea in children under five years of age (9).
2. Objectives
Given the increasing number of pediatric patients admitted to the children’s hospital in Kirkuk, especially those with UTIs who fail to respond to treatment, the study aimed to identify the most frequent specimens and investigate the epidemiology of predominant bacteria using the Multiple Antibiotic Resistance Index (MARI).
3. Methods
3.1. Sample Collection
A total of 299 samples were collected from males and females aged less than one to 15 years at the pediatric hospital in Kirkuk Governorate, during the period from May 1, 2022, to October 1, 2023.
3.2. Culturing Samples
The samples were cultured and examined under a microscope using the API20E and API Staph systems after being incubated for 24 hours at 37°C on blood agar, mannitol salt agar, and MacConkey agar.
3.3. Bacterial Sensitivity Study
Bacterial sensitivity was determined using the Kirby-Bauer disk diffusion method, where the inhibition zone was measured using a ruler (in millimeters).
3.4. Multiple Antibiotic Resistance Index
Multiple Antibiotic Resistance Index was calculated and interpreted as described by Krumperman (10), applying the formula a/b, where "b" represents the total number of antibiotics tested and "a" represents the number of antibiotics to which a single isolate is resistant (11).
3.5. Ethical Approval
Ethical approval was obtained from the Scientific Research Ethics Committee of the Ministry of Health - Kirkuk Health Department under Order 1263, dated January 2022.
4. Results
A total of 299 samples were collected from suspected patients, as diagnosed by the examining physician, at the pediatric hospital in Kirkuk Governorate, between May 1, 2022, and October 1, 2023. As shown in Table 1, most of the samples were urine, with a total of 221 samples, of which 66 (28.50%) showed positive growth and 155 (71.49%) showed negative growth. Blood samples followed, with 51 samples, 4 (9.61%) of which were positive growth and 47 (90.38%) were negative growth. Stool samples accounted for 13 samples, 2 (15.38%) of which were positive growth and 11 (84.61%) were negative growth. The numbers of Cerebrospinal fluid (CSF), throat swab, and vaginal swab samples were 1, 2, and 11, respectively, with no bacterial growth observed in any of them (Table 2).
| Isolation Source | Total Number | Bacterial Growth a | |
|---|---|---|---|
| Positive Growth | Negative Growth | ||
| Urine | 221 | 66 (28.50) | 155 (71.49) |
| Blood | 51 | 4 (9.61) | 47 (90.38) |
| Stool | 13 | 2 (15.38) | 11 (84.61) |
| CSF | 11 | - | 11 (100) |
| Throat swab | 2 | - | 2 (100) |
| Vaginal swab | 1 | - | 1 (100) |
Percentage of Bacterial Growth According to Sources of Isolation
| Source of Isolation (No.) and Bacterial Species | No. (%) |
|---|---|
| Urine (66) | |
| Escherichia coli | 28 (42.42) |
| Klebsiella pneumoniae | 13 (19.696) |
| Staphylococcus spp. | 12 (19.047) |
| Enterobacter spp. | 7 (10.606) |
| Pseudomonas spp. | 4 (6.06) |
| Proteus spp. | 1 (1.515) |
| Streptococcus spp. | 1 (1.515) |
| No identification | 6 (9.09) |
| Blood (4) | |
| Acinitobacter spp. | 1 (25) |
| Enterobacter spp. | 1 (25) |
| Pseudomonas spp. | 1 (25) |
| Klebsiella spp. | 1 (25) |
| Stool (2) | |
| E. coli | 2 (100) |
| CSF (0) | - |
| Throat swab (0) | - |
| Vaginal swab (0) | - |
Percentage of Isolates According to Sources of Isolation
4.1. Resistance of the Most Isolated Bacteria from Urine to Antibiotics
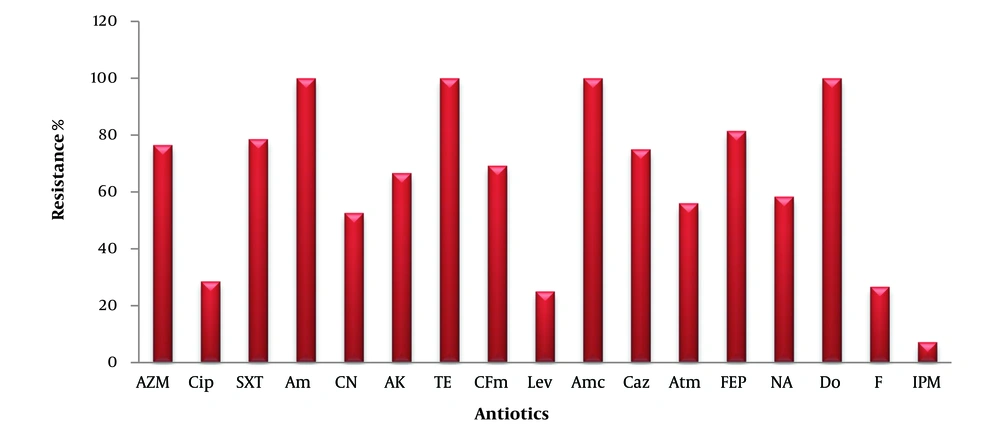
Figure 1 shows the antibiotic resistance profile of E. coli bacteria isolated from urine. The bacteria exhibited 100% resistance to the antibiotics ampicillin (Am), tetracycline (Te), amoxicillin/clavulanic acid (AMC), and doxycycline (Do). The resistance percentages for the following antibiotics were as follows: Azithromycin (AZM) (76.47%), ciprofloxacin (CIP) (28.57%), trimethoprim-sulfamethoxazole (SXT) (78.57%), cephalexin (CN) (52.63%), amikacin (AK) (66.66%), cefixime (CFM) (69.23%), levofloxacin (LEV) (25%), ceftazidime (CAZ) (75%), aztreonam (ATM) (56%), cefepime (FEP) (81.48%), nalidixic acid (NA) (58.33%), nitrofurantoin (F) (26.66%), and imipenem (IPM) (7.14%).
Resistance to antibiotics in Escherichia coli isolated from urine. AZM, azithromycin; CIP, ciprofloxacin; SXT, trimethoprim-sulfamethoxazole; AM, ampicillin; CN, cephalexin; AK, amikacin; TE, tetracycline; CFM, cefixime; LEV, levofloxacin; AMC, amoxicillin-clavulanate; CAZ, ceftazidime; ATM, aztreonam; FEP, cefepime; NA, nalidixic acid; DO, doxycycline; F, nitrofurantoin; IPM, imipenem.
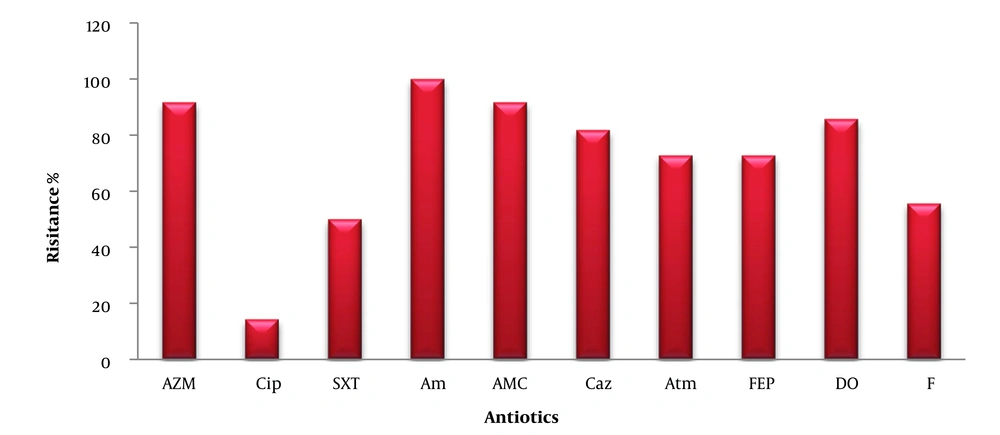
The bacteria showed resistance to AZM, CIP, SXT, AM, AMC, CAZ, ATM, FEP, DO, and F, with resistance percentages of 91.66%, 14.28%, 5%, 100%, 91.66%, 81.81%, 72.72%, 72.72%, 85.71%, and 55.55%, respectively (Figure 2).
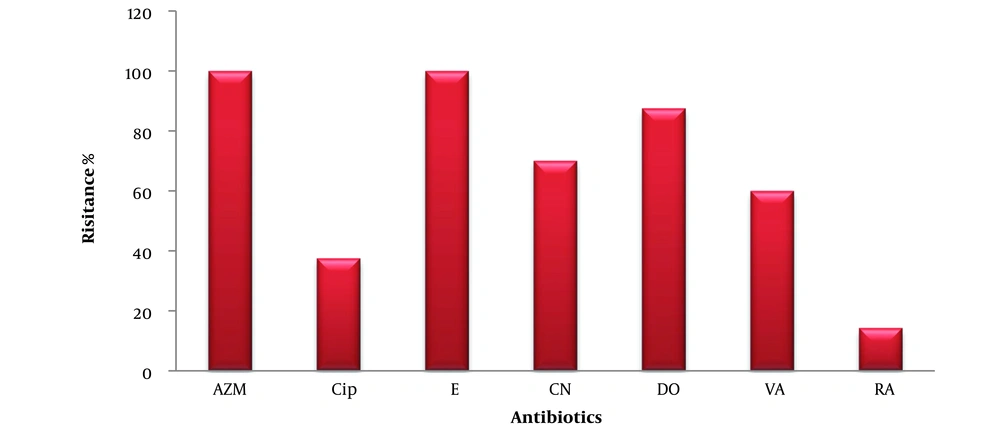
The isolates showed 100% resistance to erythromycin (E) and AZM, and resistance to CIP, CN, DO, vancomycin (VA), and rifampin (RA) at percentages of 37.5%, 70%, 87.5%, 60%, and 14.28%, respectively (Figure 3).
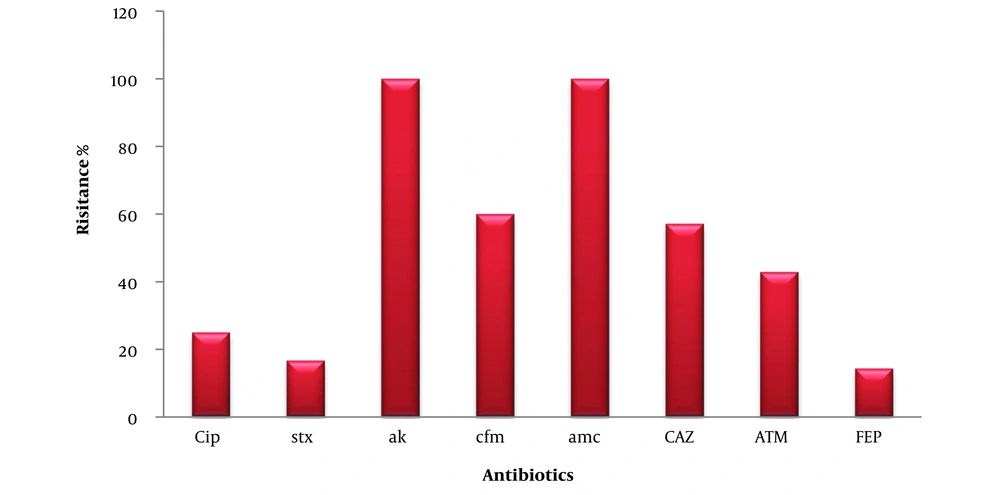
The isolates exhibited 100% resistance to AK and AMC, and resistance to CIP, SXT, CFM, CAZ, ATM, and cefepime (FEP) at percentages of 25%, 16.66%, 60%, 57.14%, 42.85%, and 14.28%, respectively (Figure 4).
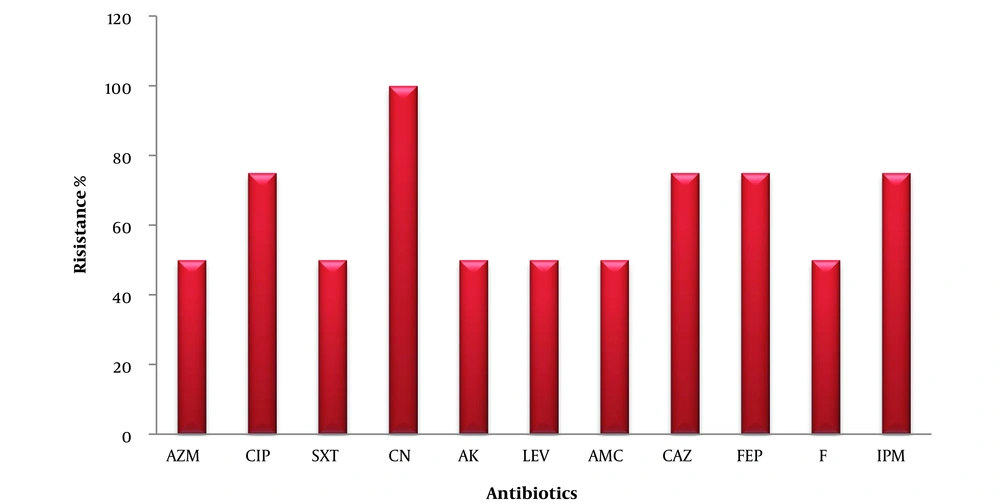
The isolates were resistant to CN by 100%, CIP, CAZ, FEP, and IPM by 75%, and AZM, SXT, AK, LEV, and F by 50% (Figure 5).
Enterobacter species isolated from urine that are resistant to antibiotics. AZM, azithromycin; CIP, ciprofloxacin; SXT, trimethoprim-sulfamethoxazole; CN, cephalexin; AK, amikacin; LEV, levofloxacin; AMC, amoxicillin-clavulanate; CAZ, ceftazidime; FEP, cefepime; F, nitrofurantoin; IPM, imipenem.
4.2. Multiple Antibiotic Resistance Index for Bacterial Isolates from Urine
Table 3 shows the MARI for bacterial isolates from urine. The MARI was greater than 0.2 in 27 isolates of E. coli (96.42%), 13 isolates of Klebsiella spp. (100%), 11 isolates of Staphylococcus spp. (91.66%), 7 isolates of Enterobacter spp. (100%), and 4 isolates of Pseudomonas aeruginosa (100%).
| MARI | Escherichia coli | Klebsiella Species | Staphylococcus Species | Enterobacter Species | Pseudomonasaeruginosa |
|---|---|---|---|---|---|
| < 0.2 | 1 (3.57) | 0 (0) | 1 (8.33) | 0 (0) | 0 (0) |
| 0.2 -0.6 | 18 (64.28) | 8 (61.53) | 6 (50) | 6 (85.71) | 0 (0) |
| 0.7 - 0.9 | 7 (25) | 5 (38.46) | 3 (25) | 0 (0) | 4 (100) |
| 1 < | 2 (7.14) | 0 (0) | 2 (16.66) | 1 (14.28) | 0 (0) |
Multiple Antibiotic Resistance Index of Bacterial Isolates from Urine a
5. Discussion
Overprescription of antibiotics is common in primary healthcare facilities. To reduce the likelihood of antibiotic resistance developing, health authorities should strictly regulate or outright prohibit the overprescription of antibiotics (12).
It is reported that E. coli (64.5%), Klebsiella species (11.6%), and Enterococcus species (6.1%) were the most common bacteria isolated from urine samples (13). This was supported by Luo et al. (14), which indicated that gram-negative bacteria were the most frequently isolated species in UTIs. A study highlighted the predominance of E. coli bacteria in patients suffering from UTIs (15).
The results indicated that bacteria isolated from urine samples exhibited resistance to many antibiotics, classifying them as multidrug-resistant (MDR) bacteria. This was confirmed by a study (16), who recorded the resistance of gram-negative bacteria to each of the following antibiotics: Gentamicin, Am/sulbactam, CIP, IM, FEP, LEV, ATM, and AK, with resistance rates of 69.56%, 56.52%, 43.47%, 0%, 52.17%, 82.60%, 60.87%, and 39.13%, respectively.
Similarly, Islam et al. (17) found that E. coli and Klebsiella species were resistant to a range of antibiotics, including sulfonamides (56%, 41%), fluoroquinolones (69%, 53%), third-generation cephalosporins (69%, 58%), macrolides (70%, 76%), and penicillin (85%, 95%), respectively.
According to a study (18), a significant proportion of antibiotic resistance was observed in K. pneumoniae and E. coli. K. pneumoniae demonstrated high resistance to streptomycin (88%), E (88%), cloxacillin (96%), while E. coli exhibited resistance to all three antibiotics.
The antibiotics IM (95%), STX (69.8%), ATM (60.5%), chloramphenicol (45.3%), and meropenem (27.9%) were more effective against Pseudomonas isolates (19).
Regarding the resistance of Pseudomonas bacteria, the results of a study (19) indicated that Pseudomonas isolates exhibited greater resistance to IM (95%), STX (69.8%), ATM (60.5%), chloramphenicol (45.3%), and meropenem (27.9%).
In contrast, Mapipa et al. (20) reported that the antibiotic ceftazidime had the highest resistance rate (63%) against P. aeruginosa isolates, with resistance to other antibiotics ranging between 7% and 35%.
Regarding the antibiotic resistance of Staphylococcus bacteria, a study (21) indicated that all Staphylococcus isolates were 100% resistant to AZM and E, and 95.56% resistant to cefixime, 50% resistant to Am, and 95% resistant to amoxicillin. Meanwhile, another study (22) indicated that all Staphylococcus aureus isolates were 100% resistant to Am and penicillin, 97.6% resistant to AK, and 90% resistant to ciprofloxacin and gentamicin.
According to a study (23), the development of quinolone resistance in Enterobacteriaceae is a complex and multifaceted process. The primary resistance mechanisms include one or more genetic mutations at the target site that alter the drug's affinity for binding to target enzymes, overexpression of the AcrAB-TolC MDR efflux pumps, and decreased expression of porins and plasmid-coded resistance proteins, such as the protection protein Qnr.
A study assessed gene sequencing in E. coli bacteria and indicated the presence of the genes orf00490, orf00819, orf001916, and orf01200, which regulate the expression of the enzyme fumarate reductase subunit D (frdD) as well as the cell division protein FtsI (penicillin-binding protein 3) (24). Additionally, the outer membrane porin protein OmpD is controlled by the genes orf00490, orf00819, and orf001916.
The occurrence of several mutations in these genes leads to bacterial resistance to many antibiotics, while the gene orf04094 expresses histidine kinase, orf02235 expresses multidrug resistance, and orf03479 expresses valine-glycine repeat G, which is excreted through the type VI secretion system (T6SS), one of the extracellular substances that contribute to antibiotic resistance.
The resistance of P. aeruginosa to cephalosporins, carbapenems, aminoglycosides, and fluoroquinolones is primarily due to horizontal gene transfer, involving integrons, plasmids, and transposons (25).
It is noted from the results that the MARI of bacteria isolated from urine was mostly greater than 0.2, indicating the extent of the epidemic of each of the bacteria (E. coli, K. pneumoniae, Enterobacter spp., Pseudomonas spp.).
This is supported by Ayandele et al. (18), which found that E. coli had the highest MAR index, reaching 1.00 in 17 isolates that showed resistance to 14 antibiotics. Meanwhile, another study (26) indicated in their study that 44% of Staphylococcus spp. isolates and 50% of E. coli isolates had MAR indices greater than 0.2.
In a study, the MARI of Pseudomonas was 0.85 (27), while a study by Ayandele et al. (18) showed in their study that the MARI in Pseudomonas isolates ranged between 0.0 and 0.8.
As for Enterococcus bacteria, a study (28), indicated in their study that the MARI for Enterococcus ranged between 0.08 and 0.83, while another study (20) showed that the MARI for P. aeruginosa ranged between 0.23 and 0.38.
Multiple Antibiotic Resistance Index is a useful tool for assessing the vulnerability of humans to the risk of resistant bacteria, as well as the extent of environmental danger due to antibiotic-resistant bacteria (10). This is especially important because antibiotic-resistant environmental bacteria can evolve into pathogens through genetic association (29) and phenotypic diversity between environmental and clinical bacteria (30). Additionally another study (31) indicated that a rise in MARI to 0.82, 0.73, and 0.64 signifies significant contamination with Vibrio parahaemolyticus bacteria. The MARI for Pseudomonas putida isolated from fish was 0.76, indicating high antibiotic use in fish farms (32). The rise of MARI above 0.2 signifies the extensive use of antibiotics in the aquatic environment. This variation in the MARI of isolated bacteria is attributed to mobile genetic elements, particularly Class 1 integrons, which provide a significant opportunity for the spread of antibiotic resistance among fish.
While Mishra et al. (33) indicated that Enterobacter with an MARI above 0.3 was observed to possess outer membrane proteins as well as other virulence factors, such as Type 1 fimbriae, biofilm production, and serum resistance.
This is supported by a study (34), which noted that there is a difference in MARI between bacterial genera. Pseudomonas aeruginosa had a higher MARI than Klebsiella spp. and Proteus spp., which in turn had a higher MARI than Enterobacter spp. The study also pointed out that 25% of the isolates posed a significant risk to humans and animals, with the majority being P. aeruginosa, which poses a higher threat as a potential pathogen compared to Enterobacter spp., based on MARI and virulence factors.
It was indicated that bacterial isolates with an MARI greater than 0.3 and a virulence factor above 0.5 pose a significant threat, while isolates with an MARI less than 0.3 and a virulence factor less than 0.5 represent a medium risk (34). Isolates with low risk have both an MARI less than or equal to 0.3 and a virulence factor greater than or equal to 0.5. Non-risk isolates are those with an MARI greater than 0.3 and a virulence factor greater than 0.5.
It is noted from the results that the values of MARI vary between genera as well as between the same bacterial species. This may be due to many factors, as indicated by (35-37), who found that differences in MARI values depend on the source of the sample, geographical location, and the method of testing.
5.1. Conclusions
According to the current study, UTIs are the most common pathological cases among young people, and urine is one of the most frequently examined samples. Escherichia coli is the most common bacterium, and antibiotic-resistant bacteria spread through the increasing MARI of isolated strains, including Enterobacter species, Pseudomonas species, E. coli, and K. pneumoniae.