1. Background
Staphylococcus aureus, one of the most prevalent hospital pathogens, poses a significant public health challenge due to its growing resistance to most antibiotics (1). Vancomycin, a glycopeptide antibiotic produced by Streptomyces orientalis, has a significant inhibitory effect on gram-positive bacteria, particularly S. aureus (2). This antibiotic is the preferred treatment for infections caused by methicillin-resistant S. aureus (MRSA), multi-drug-resistant S. aureus, and for patients with allergic reactions to semisynthetic penicillin or cephalosporins (3).
However, several recent investigations have reported increasing resistance rates to vancomycin in S. aureus clinical isolates (3, 4). The growing spread of antibiotic resistance and limited treatment options present critical challenges in managing S. aureus-related infections (5). Consequently, with the emergence of vancomycin-intermediate-resistant S. aureus (VISA) and vancomycin-resistant S. aureus (VRSA) strains, it has become essential to explore effective alternatives or potent, safe antimicrobial agents to address this problem (2-4).
Antimicrobial peptides (AMPs), expressed across various species, are well known for their broad-spectrum efficacy against bacteria, fungi, viruses, and parasites (6). In our previous research, a novel antimicrobial peptide, dendrocin-ZM1 (DZM1), was successfully identified from Zataria multiflora Boiss. This peptide, consisting of 33 amino acids, exhibits a net charge of +7, 54% hydrophobicity, amphipathic alpha-helical conformation, and negligible cytotoxicity to the HEK293 cell line. Dendrocin-ZM1demonstrated remarkable antimicrobial activity against both gram-negative and gram-positive bacterial strains, with a minimum inhibitory concentration (MIC) of 16 μg/mL against VRSA (7).
Previous studies have shown that AMPs combined with conventional antibiotics can produce a synergistic antimicrobial effect (8-12). This combination strategy has gained significant importance in various fields, especially medicine and pharmaceuticals, as a promising approach to combating resistant pathogens (11, 12).
2. Objectives
Building on previous research, our objective was to evaluate the potential of DZM1 to enhance the efficacy of vancomycin in combating various MRSA strains. Initially, we assessed the effectiveness of DZM1 and vancomycin against different MRSA strains, including a clinical isolate and MRSA ATCC 43300. Subsequently, we investigated whether subinhibitory concentrations of DZM1 could potentiate the activity of vancomycin against these MRSA strains. These findings aim to provide valuable insights into the synergistic mechanisms between antimicrobial peptides and conventional antibiotics, offering promising strategies to address the challenges of bacterial resistance.
3. Methods
3.1. Bacterial Strains
MRSA ATCC 43300, mecA-positive MRSA strains, and five MRSA clinical strains isolated from specimens provided by the Department of Microbiology at Shahid Beheshti University of Medical Sciences (Tehran, Iran) were selected for this research. Ethical approval for the study was obtained from Shahid Beheshti University of Medical Sciences, Tehran, Iran (IR.SBMU.RETECH.REC.1398.349). The presence of mecA DNA in methicillin-resistant isolates was confirmed using PCR. A few colonies were selected and suspended in 200 μL of lysis buffer. Following incubation at 30°C for 45 minutes, the mixture was boiled for 5 minutes and then diluted with 400 μL of TE buffer. For the PCR reaction, 1 μL of the lysate was added to 24 μL of the reaction mixture. The primers used to amplify the mecA gene were 5′-GTT GTA GTT GTC GGG TTT GG-3′ and 5′-CTT CCA CAT ACC ATC TTC TTT AAC-3′ (13).
The PCR protocol consisted of 30 cycles, with each cycle including 1 minute at 95°C, 1 minute at an annealing temperature that decreased incrementally from 65°C to 55°C during the first 10 cycles, and 1 minute at 72°C, performed in a thermocycler. The resulting PCR product was analyzed on a 1.5% agarose gel stained with ethidium bromide and visualized under a UV transilluminator.
For antimicrobial activity assays, Mueller–Hinton broth (MHB) and Mueller–Hinton agar (MHA) plates (HiMedia Laboratories Pvt. Ltd., Mumbai, India) were prepared. A colony-counting assay was performed after bacterial cultures were incubated for 18 hours at 37°C in MHB or on MHA plates (13).
3.2. Antimicrobial Peptide and Antibiotics
The antimicrobial peptide used in this research was DZM1 (TTLRLNTLAYKVAWLVNVKAFWAAGRA LKKVGR), a 33-amino acid peptide synthesized using the 9-fluorenylmethoxycarbonyl (Fmoc) solid-phase peptide synthesis method. The identity of the peptide was confirmed via electrospray mass spectrometry, and its purity (greater than 95%) was validated using high-performance liquid chromatography (HPLC).
3.3. Antibacterial Activity
3.3.1. Minimum Inhibitory Concentration Assay
According to the Clinical and Laboratory Standards Institute (CLSI) guidelines, the microbroth dilution technique was employed to determine the MICs of DZM1 and antibiotics, including ciprofloxacin, clindamycin, erythromycin, fusidic acid, gentamicin, nitrofurantoin, rifampin, tetracycline, vancomycin, and mupirocin, against the MRSA ATCC 43300 strain and MRSA clinical isolates (14). The antibiotics were obtained from Sigma-Aldrich (USA) and dissolved in PBS (pH 7.4).
Briefly, bacterial strains were incubated in tryptic soy broth (TSB) overnight at 37°C. The cultures were regrown to the mid-logarithmic phase and subsequently diluted to a final concentration of 5 × 10⁵ CFU/mL. A 1 µL aliquot of peptide or antibiotics was added to each well at final concentrations ranging from 0 to 128 µg/mL for peptides and 0 to 1024 µg/mL for antibiotics. Subsequently, 99 µL of diluted bacterial cells were added to each well. The plate was incubated at 37°C for 18 hours, and turbidity was measured at an optical density of 600 nm (OD600) using a microplate reader. The MIC was determined as the lowest concentration of peptide or antibiotic that inhibited 90% of visible bacterial growth.
Pure broths containing inoculum suspensions served as positive controls, while pure broths without peptides or bacteria were used as negative controls. The entire process was repeated independently three times to ensure reliability.
3.3.2. Minimum Bactericidal Concentration Assay
To determine the minimum concentration of peptide and antibiotics required to kill bacteria, a volume of 20 μL from each well showing no visible bacterial growth was plated onto Mueller-Hinton agar. The plates were incubated overnight at 37°C. The minimum bactericidal concentration (MBC) was defined as the lowest concentration at which no bacterial colonies were observed following incubation (7).
3.4. Antimicrobial Synergy Assay
The synergistic activity of DZM1 (compound B) in combination with antibiotics (compound A) was evaluated using the checkerboard broth microdilution method against MRSA strains, as previously described (15, 16). Briefly, 10 µL of peptide at 2-fold serial concentrations (0 - 64 µg/mL) was added to the vertical wells of a 96-well plate, followed by 10 µL of antibiotics at 2-fold serial concentrations (0 - 512 µg/mL) added to the horizontal wells. Next, 10 µL of bacterial suspension (2 × 106 CFU/mL) and 170 µL of fresh MHB were added to each well. The plate was incubated for 18 h at 37°C, and visible growth was assessed to determine the MIC value of the peptide/antibiotic combination.
Negative controls consisted of MHB without bacteria, while positive controls included bacterial suspensions without peptide or antibiotics.
The Fractional Inhibitory Concentration Index (FICI) was used to assess the synergistic antibacterial effects between the peptide and antibiotics. The FICI was calculated as the sum of the individual fractional inhibitory concentrations (FICs) for each compound using the formula:
The obtained FICIs then fall into one of the following categories based on their values: (1) synergy (FICI ≤ 0.5), (2) additivity (0.5 < FICI ≤ 1), (3) indifference (1< FICI ≤ 4), and (4) antagonism (4 < FICI). All experiments were carried out in triplicate.
3.5. Time Killing Kinetics
To validate the synergistic effects of peptide-antibiotic combinations observed in the checkerboard method, time-kill assays were conducted as previously described. A colony count-based assay was utilized to evaluate the time-kill kinetics of DZM1, vancomycin, and the DZM1/vancomycin combination against the MRSA ATCC 43300 strain (17).
Bacterial suspensions in the logarithmic growth phase were harvested by centrifugation (1,000 × g for 5 minutes) and washed twice with PBS. The resulting solution was then diluted to a final concentration of 2 × 105 CFU/mL. The bacterial suspension was treated with DZM1 and vancomycin at the minimum concentrations that demonstrated synergistic effects. These dilutions were prepared in 96-well plates. The solutions were incubated at 37°C with shaking at 120 rpm.
At specific time intervals (0, 5, 10, 20, 30, 60, 120, and 180 minutes), 50 µL aliquots of the mixture were withdrawn and subjected to 10-fold serial dilutions in MHB. The dilutions were subsequently plated on MHB agar plates. After overnight incubation, colony counts were performed. Each experiment was carried out in triplicate to ensure reliability.
3.6. Transmission Electron Microscopy
The TEM technique was employed to observe the structural alterations of the MRSA ATCC 43300 strain exposed to DZM1 alone or in combination with vancomycin, as previously described with some modifications (18). A bacterial suspension (1 × 106 CFU/mL) was incubated with and without DZM1 (1/16 MBC), vancomycin (1/8 MBC), and the combination of DZM1 and vancomycin (1/16 MBC DZM1 + 1/8 MBC vancomycin) for 6 hours at 37°C.
After incubation, the bacterial suspension was centrifuged and washed three times with PBS. The pellet was then fixed with a 2.5% (v/v) glutaraldehyde solution for 3 hours at 4°C. Following the fixation step, the pellet was washed with PBS and post-fixed in a 1% osmium tetroxide solution in PBS for 70 minutes. After two additional PBS washes, the specimens were gradually dehydrated using a series of acetone solutions and embedded in Epon 812 resin.
The embedded samples were sectioned using an ultramicrotome and stained with uranyl acetate and lead citrate. Finally, the specimens were examined using a Zeiss EM-900 TEM apparatus, operated at 80 kV.
3.7. Scanning Electron Microscopy
To investigate the morphological changes in the MRSA ATCC 43300 strain (1 × 106 CFU/mL) treated with DZM1 alone or in combination with vancomycin, SEM analysis was performed following the method described by Lyu et al. (19). The bacterial suspension was treated in the presence and absence of DZM1 (1/16 MBC), vancomycin (1/8 MBC), and a combination of DZM1 and vancomycin (1/16 MBC DZM1 + 1/8 MBC vancomycin) for 6 hours.
After 5 minutes of centrifugation, the resulting pellet was fixed with 2.5% (v/v) glutaraldehyde for 3 hours at 4°C. Following three washes with 0.1% PBS, the samples were treated with 1% osmium tetroxide in PBS for 1 hour at room temperature. The fixed samples were then washed with PBS and sequentially dehydrated using an ethanol series (25%, 50%, 75%, 95%, and 100%) for 10 minutes each, followed by treatment with absolute alcohol for 45 minutes.
The dehydrated samples were subjected to a critical-point drying technique using CO2 and subsequently coated with a thin layer (20 - 30 nm) of gold-palladium. Observations were performed using an analytical SEM microscope, the JEOL JSM-6510LA. Bacterial cells grown without the peptide and antibiotic were processed using the same protocol to serve as the control.
3.8. Statistical Analysis
All statistical analyses were conducted using GraphPad Prism version 8.01 (GraphPad Software, San Diego, CA, United States). Each test was performed in triplicate to ensure reliability and reproducibility of the results.
4. Results
4.1. Minimal Inhibitory Concentration and Minimum Bactericidal Concentration of Peptide and Antibiotics
The minimum inhibitory concentration and MBC for DZM1 against MRSA ATCC 43300 were determined to be 16 µg/mL. Dendrocin-ZM1 exhibited the same levels of activity against MRSA clinical isolates (Table 1). Overall, DZM1 demonstrated strong antimicrobial activity against MRSA ATCC 43300 and MRSA clinical isolates in this study. The microbroth dilution method revealed a high level of resistance among MRSA clinical strains to multiple antibiotics, with resistance rates of 80% for tetracycline, erythromycin, and ciprofloxacin, and 60% for clindamycin, gentamicin, nitrofurantoin, and rifampin. All isolates were found to be susceptible to vancomycin, while resistance to mupirocin was observed in all clinical MRSA isolates. In this study, all MRSA clinical isolates were confirmed as multidrug-resistant (MDR) strains but remained susceptible to vancomycin (Table 1). The data also showed that the MIC value of DZM1 against MRSA clinical isolates was consistent, except for IR3-MRSA. Additionally, the results of in vitro experiments indicated that the MBC value for all clinical isolates was higher than that for ATCC 43300.
| Strain | MIC/MBC (μg/mL) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Dendrocin‑ZM1 | VAN | TET | ERY | CLI | GEN | RIF | CIP | NIT | MUP | |
| S. aureus ATCC 43300 | 16/16 | 0.125/0.5 | 0.5/1 | 0.5/1 | 1/2 | 0.5/0.5 | 0.25/1 | 0.25/0.5 | 4/16 | 8/16 |
| IR1-MRSA | 16/32 | 1/2 | 64/64 | 8/8 | 4/4 | 32/32 | 4/4 | 4/4 | 16/16 | 256/256 |
| IR2-MRSA | 16/32 | 1/2 | 32/128 | 8/8 | 4/8 | 4/8 | 1/2 | 1/2 | 128/256 | 32/64 |
| IR3-MRSA | 32/32 | 2/4 | 16/32 | 0.5/1 | 0.5/1 | 16/32 | 0.5/1 | 8/16 | 8/8 | 512/1024 |
| IR4-MRSA | 16/32 | 1/2 | 16/32 | 16/16 | 8/8 | 2/4 | 4/4 | 8/16 | 256/256 | 256/512 |
| IR5-MRSA | 16/32 | 2/4 | 4/4 | 8/16 | 0.5/1 | 128/128 | 8/16 | 16/16 | 128/128 | 256/512 |
The Minimal Inhibitory Concentration and Minimum Bactericidal Concentration of Dendrocin-ZM1 and Antibiotics Against Methicillin-Resistant Staphylococcus aureus Strains
4.2. Antibacterial Effect of Peptide Combined with Antibiotics
Dendrocin-ZM1 demonstrated increased effectiveness when combined with antibiotics. According to our findings, antibacterial agents that were inactive when tested alone exhibited activity when combined with DZM1 (Table 2). The analysis revealed a significant synergistic effect with the DZM1/vancomycin (VAN) combination and a complete additive effect in the combinations of DZM1/erythromycin (ERY), DZM1/ciprofloxacin (CIP), DZM1/tetracycline (TET), and DZM1/mupirocin (MUP). Additionally, an additive effect was observed in 20% of the cases for DZM1/rifampin (RIF) and 80% of the cases for DZM1/gentamicin (GEN) and DZM1/clindamycin (CLI). However, DZM1 was found to have no effect on nitrofurantoin (NIT) activity, while an indifferent effect was recorded in 80% of the cases for DZM1/RIF and 20% of the cases for DZM1/GEN and DZM1/CLI. Notably, no antagonistic effects were observed in any of the cases (Table 3).
| MIC of Combination (μg/mL) | MRSA Clinical Isolates | ||||
|---|---|---|---|---|---|
| IR1-MRSA | IR2-MRSA | IR3-MRSA | IR4-MRSA | IR5-MRSA | |
| Dendrocin‑ZM1/vancomycin | 1/0.25 | 1/0.25 | 4/0.125 | 1/0.25 | 2/0.5 |
| Dendrocin‑ZM1/tetracycline | 8/16 | 4/16 | 16/8 | 4/8 | 8/2 |
| Dendrocin‑ZM1/erythromycin | 8/2 | 8/2 | 4/0.25 | 8/4 | 8/2 |
| Dendrocin‑ZM1/clindamycin | 4/2 | 4/2 | 16/0.25 | 4/4 | 16/1 |
| Dendrocin‑ZM1/gentamicin | 8/8 | 8/2 | 8/8 | 8/1 | 16/64 |
| Dendrocin‑ZM1/rifampin | 32/8 | 32/2 | 64/1 | 4/2 | 32/8 |
| Dendrocin‑ZM1/ciprofloxacin | 8/2 | 8/0.5 | 16/1 | 8/1 | 8/8 |
| Dendrocin‑ZM1/nitrofurantoin | 16/8 | 16/64 | 32/1 | 16/128 | 16/16 |
| Dendrocin‑ZM1/mupirocin | 8/128 | 8/16 | 16/256 | 8/128 | 8/128 |
Combinatory Inhibitory Effects of Dendrocin-ZM1 with Antibiotics Against 5 Methicillin-Resistant Staphylococcus aureus Strains
| peptide and Antibiotic Combinations | FICI (S, Ad, I, An) | ||||
|---|---|---|---|---|---|
| IR1-MRSA | IR2-MRSA | IR3-MRSA | IR4-MRSA | IR5-MRSA | |
| DZM1+VAN | 0.31 (S) | 0.31 (S) | 0.19 (S) | 0.31 (S) | 0.38 (S) |
| DZM1+TET | 0.75 (Ad) | 0.75 (Ad) | 1 (Ad) | 0.75 (Ad) | 1 (Ad) |
| DZM1+ERY | 0.75 (Ad) | 0.75 (Ad) | 0.63 (Ad) | 0.75 (Ad) | 0.75 (Ad) |
| DZM1+CLI | 0.75 (Ad) | 0.75 (Ad) | 1 (Ad) | 0.75 (Ad) | 3 (I) |
| DZM1+GEN | 0.75 (Ad) | 1 (Ad) | 0.75 (Ad) | 1 (Ad) | 1.5 (I) |
| DZM1+RIF | 4 (I) | 4 (I) | 4 (I) | 0.75 (Ad) | 3 (I) |
| DZM1+CIP | 1 (Ad) | 1 (Ad) | 0.63 (Ad) | 0.63 (Ad) | 1 (Ad) |
| DZM1+NIT | 1.5 (I) | 1.5 (I) | 1.12 (I) | 1.5 (I) | 1.12 (I) |
| DZM1+MUP | 1 (Ad) | 1 (Ad) | 1 (Ad) | 1 (Ad) | 1 (Ad) |
In vitro Activities of Dendrocin-ZM1 in Combination with Antibiotics Against Methicillin-resistant Staphylococcus aureus Clinical Isolates a
Given the complete synergistic effect of DZM1 with vancomycin against MRSA clinical isolates, further analysis was conducted on the VAN/DZM1 combination using the MRSA ATCC 43300 reference strain. Although DZM1 and vancomycin individually demonstrated antibacterial activity against MRSA ATCC 43300, the VAN/DZM1 combination exhibited the most pronounced response, with the MIC value of vancomycin decreasing from 1 μg/mL to 0.125 μg/mL. Similarly, the MIC of DZM1 dropped significantly from 16 μg/mL to 1 μg/mL when combined with vancomycin against MRSA ATCC 43300. A comparable trend was also observed for the MIC values of DZM1 and vancomycin against clinical isolates.
4.3. Time-Killing Kinetics of Peptide Combination with Vancomycin
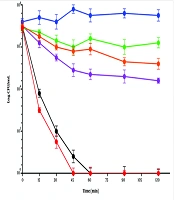
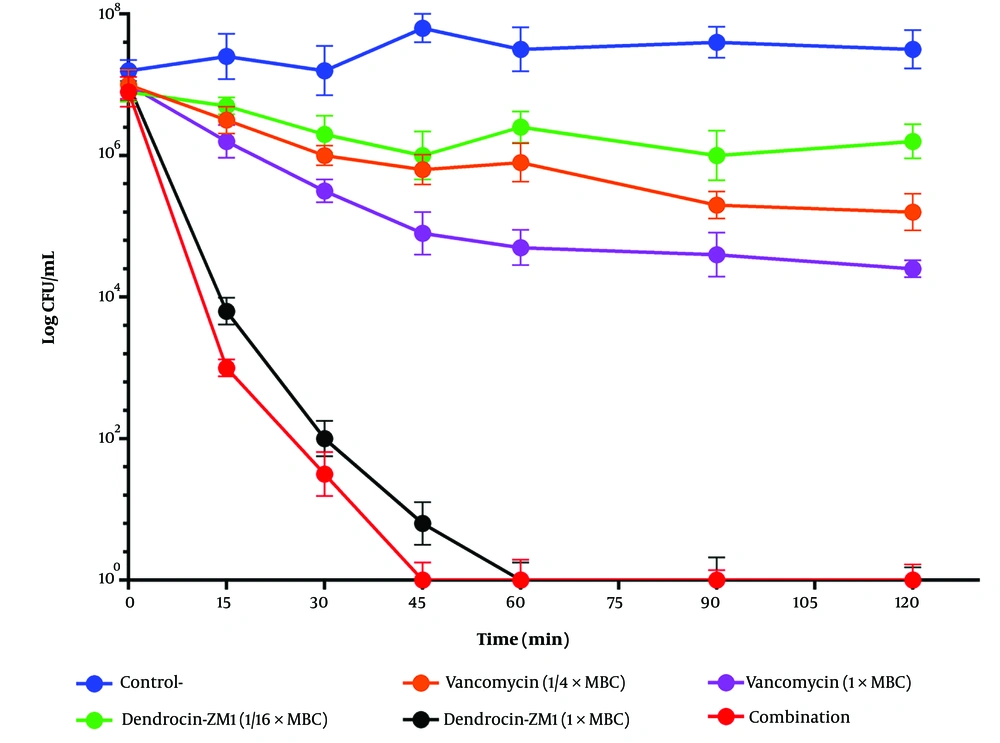
The time-killing assay was performed to evaluate the bactericidal efficacy of DZM1 in combination with vancomycin against MRSA ATCC 43300. At sub-MBC levels, neither DZM1 nor vancomycin alone exhibited significant bactericidal activity within 180 minutes against S. aureus. However, the bacteria were completely eliminated within 45 minutes when the two agents were used in combination (Figure 1). These findings indicate that the combination of DZM1 and vancomycin at sub-MBC concentrations maintained a sustained antibacterial effect against the MRSA ATCC 43300 strain.
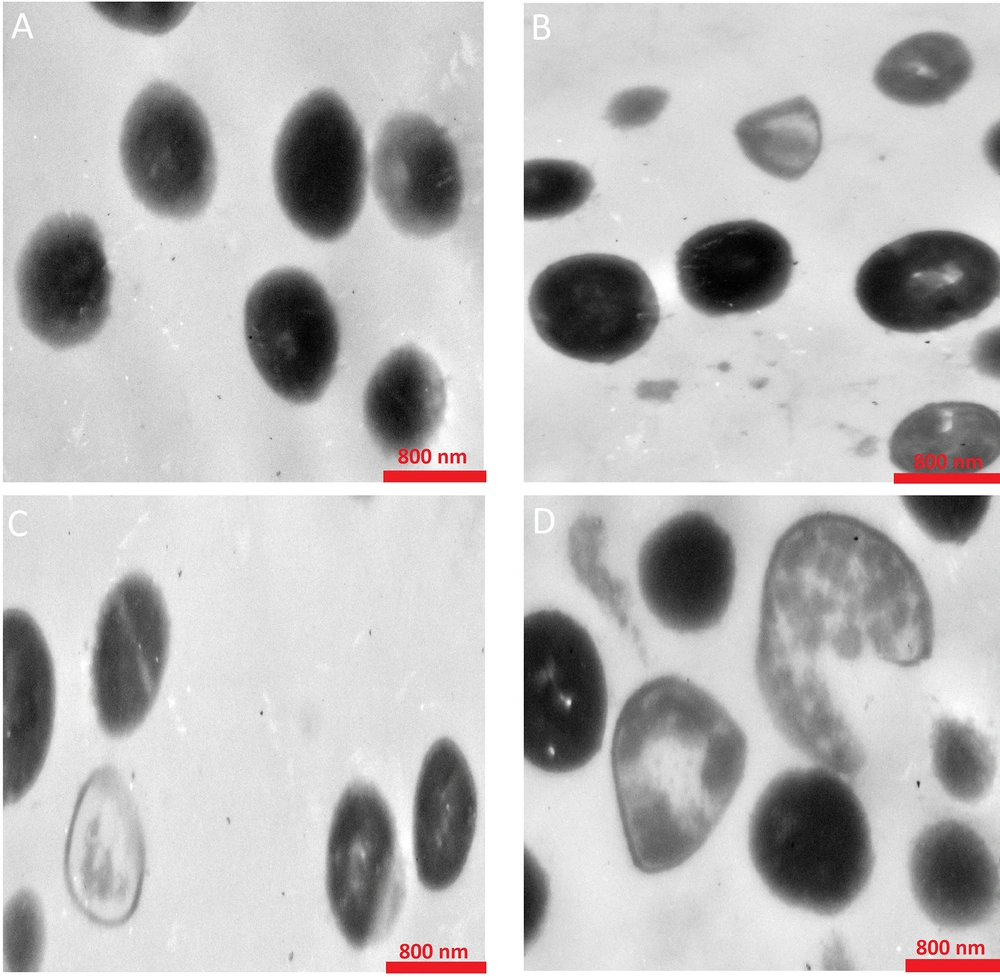
A comprehensive analysis of the morphological changes and internal structure of MRSA ATCC 43300 after treatment with the combination of DZM1 and vancomycin was conducted. As illustrated in Figure 2, TEM micrographs of MRSA ATCC 43300 in the absence of DZM1 or vancomycin revealed round, smooth cells with a bright appearance and intact cell walls and membranes. In contrast, treatment with a sub-MBC of DZM1/VAN resulted in significant cell membrane disruption and substantial leakage of cellular contents. Notably, the cell membranes of bacteria exposed to the DZM1/VAN combination were markedly more compromised compared to those treated with DZM1 or vancomycin alone (Figure 2).
Transmission electron microscopy of MRSA ATCC 43300 cells treated with Dendrocin‑ZM1, vancomycin and DZM1-vancomycin combinations. A, control or untreated MRSA About 1 × 106 bacterial cells were incubated; with B, vancomycin (1/8 MBC); C, Dendrocin‑ZM1 (1/16 MBC); D, DZM1+vancomycin (1/16 MBC DZM1 + 1/8 MBC vancomycin) for 3 h. Exposure to Dendrocin‑ZM1 and vancomycin resulted in morphological changes including loss of the structural integrity of the cell membrane and shrunk cytoplasm membrane detached from the cell wall and destruction of the cell wall.
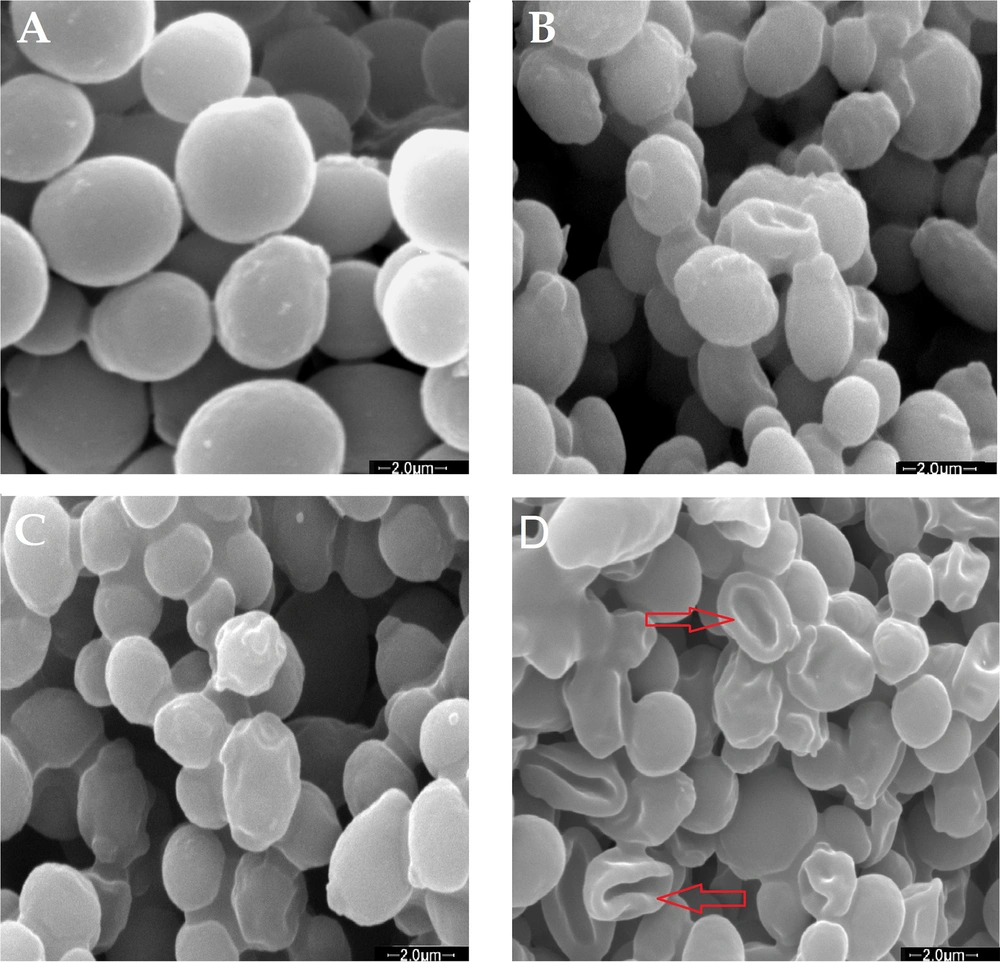
The bacterial morphology of MRSA ATCC 43300 was further analyzed using SEM following treatment with the combination of DZM1 and vancomycin. In the control samples of S. aureus, the cells appeared round, smooth, and undamaged (Figure 3A). After exposure to either DZM1 or vancomycin alone, some bacterial cells exhibited noticeable alterations, including surface wrinkles and moderate deformities (Figure 3B and C). However, the combination treatment of DZM1 and vancomycin resulted in severe cellular damage, characterized by deep wrinkles, significant deformities, and substantial leakage of cellular contents (Figure 3D). These findings demonstrate that the combination of DZM1 and vancomycin at sub-MBC levels exhibited an enhanced bactericidal effect by promoting extensive cellular damage.
Scanning electron microscopy of MRSA ATCC 43300 cells treated with Dendrocin-ZM1, vancomycin and DZM1-vancomycin combinations. A, control or untreated MRSA. About 1 ×106 bacterial cells were incubated; with B, vancomycin (1/8 MBC); C, Dendrocin-ZM1 (1/16 MBC); D, DZM1 + vancomycin (1/16 MBC DZM1 + 1/8 MBC vancomycin) for 3 h. SEM images of the untreated cells indicated the cells round and intact, (A); Cells treated with Dendrocin-ZM1 or vancomycin alone exhibited a series of slight and superficial changes in bacterial cells, (B and C); dendrocin-ZM1+ vancomycin combinations treated MRSA indicated major changes such as deformity and deep wrinkles and completely lysed cells (red arrows), (D).
5. Discussion
The rapidly increasing prevalence of MDR S. aureus strains and the lack of significant advancements in discovering new therapies pose challenges for treating staphylococcal infections. This issue is compounded by antibiotic resistance, which has become a major obstacle in infection control (1, 20). Developing antimicrobials that target S. aureus virulence factors presents a promising approach for managing S. aureus-induced infections (12). In recent decades, the prevalence of MRSA isolates resistant to multiple drugs has risen, leaving vancomycin as the only viable treatment option. However, reports of resistance to vancomycin, including the emergence of VISA and VRSA, are particularly alarming (2). These findings highlight the growing concern surrounding VRSA infections and emphasize the need for vigilant attention, as vancomycin represents the final therapeutic option for treating MRSA (21).
Epidemiological evidence underscores the potential utility of combining novel AMPs with conventional antibiotics, such as vancomycin, to effectively combat MDR-MRSA infections (1, 2, 21). Numerous studies have demonstrated the broad-spectrum antimicrobial activity of Zataria multiflora Boiss (7, 22). Consistent with previous research, our earlier study (7) revealed that a peptide purified from Zataria multiflora Boiss, named DZM1, exhibited antibacterial activity against MSSA and MRSA strains. Furthermore, we demonstrated that DZM1 displayed synergistic activity with vancomycin against MRSA strains.
In vitro experiments in the current study confirmed the potent antibacterial efficacy of DZM1 against MRSA clinical isolates and MRSA ATCC 43300, aligning with findings from earlier studies (7, 23).
According to the results of the FIC assay, we demonstrated the synergistic inhibitory effect of the DZM1+VAN combination on five clinical MRSA strains and MRSA ATCC 43300, which effectively inhibited bacterial growth. Our study revealed that combining DZM1 with vancomycin reduced the MIC of the peptide by 16-fold and vancomycin by 3-fold against MRSA. Therefore, the DZM1+VAN combination might serve as a potential option for the effective treatment of MRSA infections, similar to findings reported by other researchers. In a recent study by Roshanak et al., a synergistic effect was observed when cLFchimera was combined with vancomycin (FIC: 0.375) (24). Earlier data published by Wu et al. reported the enhanced activity of AMPs Bip-P-113 and Nal-P-113 when used in combination with vancomycin against VRSA strains (25). Several studies have also documented the synergistic inhibitory effects of AMPs combined with conventional antibiotics (8, 10, 11).
Wu et al. demonstrated that combining various AMPs with azithromycin significantly improved bactericidal efficacy against multidrug-resistant bacteria, including S. aureus (26). Similarly, Jorge et al. assessed the synergistic impact of colistin and AMPs against two major pathogens, Pseudomonas aeruginosa and S. aureus, in single- and double-species biofilm cultures (27). Our results indicated that the MIC value of DZM1 was acceptable when compared with other antibiotics. The findings clearly demonstrate that the combination of vancomycin with DZM1 is a successful approach to enhancing the efficacy of both the peptide and vancomycin against MRSA clinical isolates.
It is worth noting that reducing the quantity of antibiotics employed could help mitigate the development of resistance. Overall, the aforementioned findings indicate that the development of AMPs, either as alternatives or in combination with traditional antibiotics, holds great promise. The differences observed between DZM1 and antibiotic activities may be attributed to their distinct structures and bactericidal mechanisms. Antimicrobial peptides are known to exhibit their bactericidal activity by targeting the cell membrane rather than specific molecular targets. Consequently, they demonstrate antimicrobial action against a broad spectrum of pathogens and offer practical antibacterial tools without the tendency to promote microbial resistance (28).
The combination strategy that reduces the MIC values of DZM1 could be a valuable approach for the clinical development of AMPs. This reduction in MIC values allows AMPs to be used at lower concentrations, addressing the problem of AMP-related toxicity.
In this study, it was demonstrated that the release of cellular content was significantly higher with the use of the DZM1/VAN combination compared to when the bacteria were treated with the peptide or antibiotic alone. These findings support the hypothesis that DZM1 exhibits its bactericidal activity through a membrane disruption pathway, which may facilitate antibiotic inflow and ultimately induce cell death. These results align with those of Wu et al., who reported that the cell wall-disrupting function was dramatically enhanced in the presence of DP7 when combined with AZT or VAN, compared to the untreated control (26).
Similarly, Zhao et al. reported that MP1102 exerted its bactericidal activity by destroying cell membrane integrity and interacting with cell DNA in Streptococcus suis (29). A study by Schneider et al. revealed that CATH-2 damages the cell wall and increases cellular membrane permeability, thereby eliminating S. aureus and E. coli (30). Comparable findings have shown that nisin and oxacillin exert synergistic antibacterial effects on MRSA strains. These effects may result from their ability to damage the cell wall, alter cellular membrane permeability, and disrupt cellular integrity, ultimately leading to the release of intracellular contents and bacterial death (10).
Moreover, a study from China reported that the combination of CATH-2 and erythromycin effectively disrupted the cell membrane of E. coli at a significantly higher level compared to the individual effects of CATH-2 or erythromycin alone (9).