1. Background
Acinetobacter baumannii is an opportunistic, Gram-negative pathogen known to cause a variety of healthcare-associated infections in immunocompromised patients, including bacteremia, meningitis, pneumonia, urinary tract infections, and wound infections (1). Patients who contract these infections, particularly ventilator-associated pneumonia and bloodstream infections, face elevated risks of significant mortality rates, ranging from 5% in general hospital wards to 54% in the intensive care units (ICU). Additionally, there is an increasing incidence of community-acquired A. baumannii infections being reported (2).
The rapid emergence of A. baumannii strains resistant to nearly all available antibiotics, including β-lactams, carbapenems, fluoroquinolones, macrolides, and aminoglycosides, has further necessitated the use of colistin (also known as polymyxin E) as the only remaining treatment option (3-5). Although colistin is considered the last resort for treating drug-resistant A. baumannii, reports of clinical isolates resistant to this antibiotic have risen (6, 7). Resistance to colistin is attributed to both chromosomally-encoded resistance mechanisms and plasmid-borne mcr genes (mcr-1 to mcr-10) in various bacterial species. However, only four mobile colistin resistance genes (mcr-1 to mcr-4) have been identified in A. baumannii isolates (2, 8, 9).
Considering that the dissemination of plasmid-borne colistin resistance jeopardizes this treatment option, epidemiological surveillance assumes a crucial role in controlling and preventing the spread of the resistance mechanism (10). Up to this point, most studies on A. baumannii clinical isolates have primarily focused on the prevalence of the mcr-1 gene, with limited attention given to other mcr genes (9, 11-14).
2. Objectives
This knowledge gap could impact the development of effective treatments for A. baumannii infections. Therefore, this study aims to evaluate the prevalence of mcr genes (mcr-1 to mcr-3) in A. baumannii strains recovered from Shahid Rahimi Hospital.
3. Methods
3.1. Clinical Specimens, Bacterial Isolates, and Identification
The present cross-sectional study examined A. baumannii isolates obtained from various specimens, including blood, wound, sputum, urine, and cerebrospinal fluid (CSF) samples. These samples were collected from patients referred to Shahid Rahimi Hospital in Khorramabad, Iran, during a seven-month period, from January to August 2019. In this research, hospitalized patients with laboratory-confirmed A. baumannii infections during the specified study period were included, while those without a confirmed A. baumannii infection or whose infections were attributed to microorganisms other than A. baumannii species were excluded.
The specimens were inoculated onto blood agar and MacConkey agar plates and then incubated overnight at 37°C. The primary identification of the A. baumannii isolates was performed using conventional microbiological and biochemical methods, as previously described (15). Subsequent confirmation of A. baumannii was carried out through PCR for the blaoxa-51-like gene (15).
This study was approved by the local ethics committee of Lorestan University of Medical Sciences (IR.LUMS.REC.1399.340).
3.2. Antimicrobial Susceptibility Testing
The minimum inhibitory concentrations (MICs) of colistin (Colistin Sulfate salt powder, Sigma-Aldrich, St. Louis, MO, USA) were determined using the broth microdilution method, following CLSI guidelines (CLSI, 2023). Stock solutions were prepared from colistin in distilled water to provide concentrations ranging from 0.25 μg/mL to 256 μg/mL. Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 strains were used as quality control strains.
3.3. DNA Extraction
DNA extraction was performed using the boiling method, and the DNA samples were subsequently stored at -70°C until further use (16).
3.4. Detection of Plasmid-Mediated Colistin-Resistant Genes
Multiplex PCR with specific primers, as described in Table 1, was used to screen for the presence of mcr-1 to mcr-3 genes. The Multiplex PCR reaction was performed in a total volume of 25 μL containing 2X Taq PCR Master Mix, 10 µM of each primer, and 5 μL of template DNA. Following amplification, the resulting product was analyzed by electrophoresis on a 1.5% agarose gel. The strain provided by Prof. C. Giske from Karolinska Institute, Sweden, was used as a positive control, and sterile water was used as a negative control.
Primer Sequences Used for Multiplex PCR for Detection of mcr-1 to mcr-3 in Clinical Isolates of Acinetobacter baumannii
3.5. Statistical Analysis
Data analysis was conducted using SPSS version 26 (SPSS, Inc., Chicago, IL, USA) with Fisher's exact tests employed, and P-values less than 0.05 were considered statistically significant.
4. Results
4.1. Characteristics of Patient Demographics and Clinical Isolates
A total of 47 A. baumannii clinical isolates were collected and studied during the aforementioned period to identify underlying resistance mechanisms. Of these, 32 A. baumannii isolates (68.1%) were obtained from male patients, while 15 (31.9%) were from female patients. The age range of the patients varied from 20 to 90 years. The isolates primarily originated from patients admitted to the ICU, accounting for 34 (72.4%), followed by other hospital wards with 13 (27.6%).
In total, 14 (29.8%) strains exhibited resistance to colistin, with MICs ranging from 2 to 256 μg/mL (Table 2). Notably, within this group, 3 isolates demonstrated intermediate resistance, while 11 isolates exhibited full resistance to colistin. The MIC50 and MIC90 values for colistin were calculated as 0.25 μg/mL and 16 μg/mL, respectively. Among the A. baumannii isolates, 20 out of 47 (42.6%) were classified as multidrug-resistant (MDR), while 23 out of 47 (48.9%) demonstrated extensive drug resistance (XDR). Multidrug-resistant and XDR phenotypes were determined using the standardized definitions outlined in a previously published article, based on the disk diffusion results (17).
| MIC Range (μg/mL) | 2 | 4 | 8 | 16 | 32 | 256 |
|---|---|---|---|---|---|---|
| The number of isolates | 3 | 2 | 3 | 1 | 3 | 2 |
The Minimum Inhibitory Concentration Results of Acinetobacter baumannii Isolates Non-susceptible to Colistin
4.2. Molecular Characterization of the mcr Genes
A total of 47 A. baumannii isolates underwent multiplex PCR screening for the presence of mcr-1 to mcr-3 genes.
To assess the role of plasmid-encoded resistance genes (mcr-1, mcr-2, and mcr-3), PCR was performed using bacterial genomic DNA extracted from the patients.
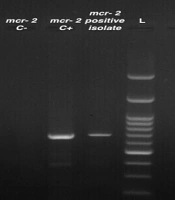
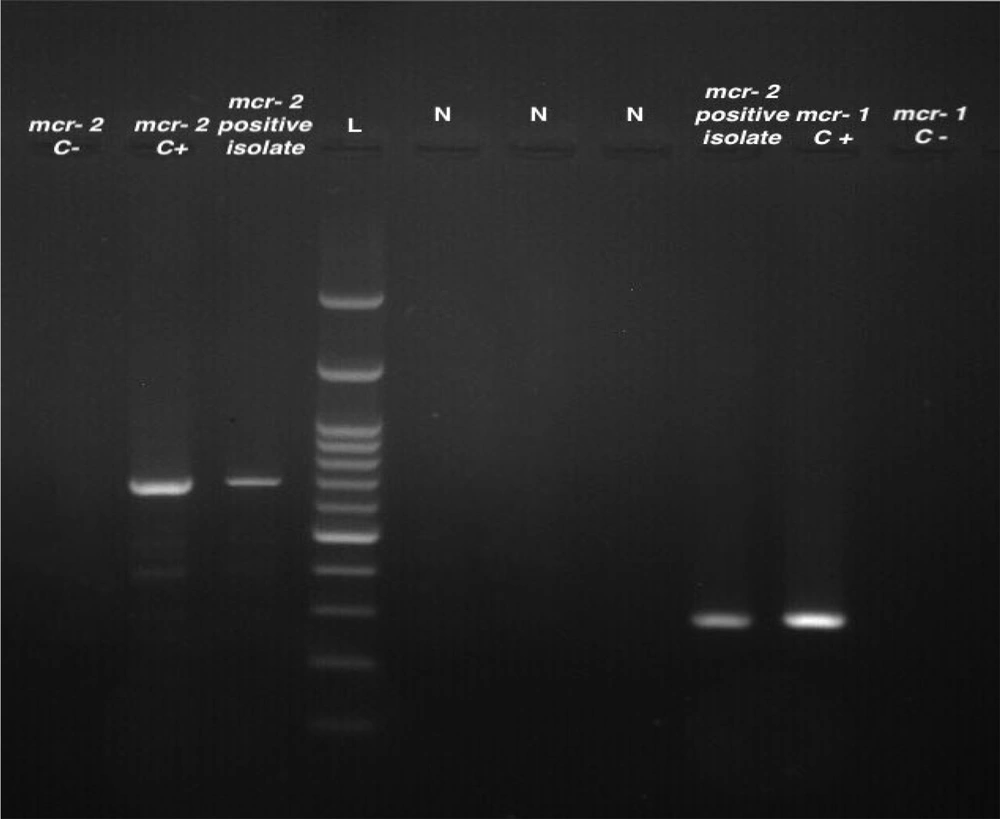
Molecular analysis of mobile colistin resistance (mcr) genes associated with A. baumannii isolates revealed that 6 (12.7%) and 2 (4.2%) of the clinical isolates were positive for mcr-1 and mcr-2, respectively. However, no isolate was positive for the mcr-3 resistance gene. Figure 1 displays the results of the PCR analysis for the detection of mcr genes in A. baumannii isolates.
Multiplex PCR products of mcr genes in A. baumannii isolates. Lane 1, mcr-2 negative control; lane 2, mcr-2 positive control; lane 3; mcr-2 positive isolate; lane 4, molecular size markers (100‐bp DNA ladder; SinaClon); lane 5 to 7, negative isolates; lane 8, mcr-2 positve isolate; lane 9, mcr-1 positive control; lane 10, mcr-1 negative control.
It should be noted that, except for one isolate, the genes responsible for colistin resistance, specifically mcr-1 and mcr-2, were detected in susceptible A. baumannii isolates with a MIC value < 2 μg/mL.
5. Discussion
Patients in ICUs face a higher risk of contracting hospital-acquired infections, including A. baumannii, compared to those in general wards. This heightened risk is primarily attributed to the frequent use of invasive devices in ICUs and increased exposure to antibiotic-resistant pathogens (18). Consequently, this study demonstrates that A. baumannii is predominantly isolated from ICU settings. Given the significant prevalence of resistance to current treatments, colistin has regained attention as a potential solution for combating severe infections caused by carbapenem-resistant A. baumannii (19, 20). However, the increased use of colistin as a treatment option has led to the emergence of colistin-resistant strains on a global scale (7).
In this study, 14 isolates (29.7%) displayed intermediate resistance or full resistance to colistin. Moreover, the emergence of colistin resistance has been reported not only in Iran but also in various parts of the world (7, 9, 21-23). Consistent with findings from recent research and similar studies conducted globally, a failure to address this issue promptly may result in frequent outbreaks of pan-drug-resistant A. baumannii strains that also exhibit resistance to colistin in the near future.
Colistin susceptibility testing is performed using broth microdilution, which is the gold standard phenotypic method, but it requires an overnight incubation to obtain results. Accordingly, the adoption of faster and more accurate techniques, such as molecular detection, is anticipated for identifying the genetic mechanisms responsible for antimicrobial resistance (24, 25). Resistance transmitted by plasmids can confer resistance to multiple antibiotics. Furthermore, plasmids can disseminate resistance among bacteria at a higher rate, potentially leading to rapid and widespread distribution in clinical settings (26, 27). As with other plasmids, plasmid-mediated mcr genes can spread horizontally among humans, animals, and the environment, posing a significant risk to human health (28).
In the current study, out of the 47 A. baumannii isolates, 6 (12.7%) and 2 (4.2%) isolates harbored mcr-1 and mcr-2 genes, respectively. However, no isolate carried the mcr-3 gene. Higher incidence rates were reported in Iraq. Al-Kadmy et al. found that out of 121 clinical isolates, 89 (73.5%), 78 (64.5%), and 82 (67.8%) of isolates were positive for the mcr-1, mcr-2, and mcr-3 genes, respectively (9). Moreover, a study by Hafudh et al. revealed that among 13 isolates with reduced sensitivity to colistin, 5 isolates (38.5%) were positive for the mcr-1 gene, and 4 isolates were positive for both mcr-2 and mcr-3 genes, respectively (29). In a study conducted by Kareem, the mcr-1 gene was detected in 22 isolates (11%), while no isolates carried the mcr-2 or mcr-3 genes (30). However, other studies in Iran and South Africa reported that none of the identified A. baumannii isolates carried the mcr resistance genes (14). For instance, studies conducted by Babaei et al. and Tehrani et al. in Tehran and Ghazvin cities revealed no detection of the mcr-1 gene among the examined A. baumannii isolates, indicating a potential regional absence of this specific colistin resistance mechanism (31, 32).
It is important to note that, with the exception of one isolate, none of the mcr-positive isolates were phenotypically resistant to colistin. This lack of resistance may be associated with the non-expression of resistance genes due to various factors (33, 34). Moreover, the prevalence of the resistance genes did not align with the number of resistant isolates detected by the phenotypic test. The lower detection rate of mcr genes compared to the phenotypically observed resistance suggests that phenotypically resistant isolates may be associated with other resistance mechanisms (7).
5.1. Conclusions
In conclusion, this study provides valuable insights into the prevalence of colistin resistance and the presence of mcr-1 and mcr-2 genes in A. baumannii clinical isolates. Given that phenotypic resistance was higher than the number of genotypically positive isolates and was largely unrelated to the presence of mcr genes, it is essential to investigate other colistin resistance mechanisms, considering its critical role as a last-resort treatment for drug-resistant A. baumannii.