1. Background
Ciprofloxacin, a quinolone antibiotic, is a broad-spectrum antimicrobial agent widely used in humans. This class of antibiotics was introduced into clinical practice in the 1960s. However, resistance rates to these agents in Enterobacteriaceae have increased globally in recent years (1). Resistance to this broad-spectrum antibiotic, which is used to treat systemic infections and chronic urinary infections caused by gram-negative bacteria such as Klebsiellapneumoniae and gram-positive bacteria, is a significant concern in the discussion of microbial drug resistance (2).
Ciprofloxacin resistance generally results from mutations in the quinolone resistance-determining region (QRDR), which encodes genes for topoisomerase IV (parC, parE) and DNA gyrase (gyrA, gyrB) (1, 3). A plasmid-mediated quinolone resistance (PMQR) mechanism was later identified as an alternative resistance mechanism. The PMQR was first described in 1998 in an isolate of K.pneumoniae (4). The PMQR mechanisms are classified into three types: (1) Proteins and enzymes that protect bacterial topoisomerases from quinolones (qnrA, qnrB, qnrC, qnrD, and qnrS), (2) acetylation enzymes such as aac(6)-Ib, which increase resistance to aminoglycosides, and (3) multidrug efflux pumps (qepA and oqxAB), which expel antibiotics from bacterial cells. These factors collectively contribute to bacterial drug resistance (4, 5).
Mobile genetic elements, typically carried by plasmids and transferred through conjugation, play a crucial role in the dissemination of antibiotic resistance genes and the development of multidrug resistance (MDR) in bacteria. This poses significant challenges in infectious disease management and public health. Additionally, integrons can capture gene cassettes containing PMQR genes from other bacteria or environmental sources and incorporate them into their genetic structure, enabling bacteria to acquire quinolone resistance genes. The horizontal transfer of integrons carrying PMQR gene cassettes facilitates the spread of quinolone resistance among bacterial populations (6).
To date, no comprehensive study has examined the prevalence of PMQR determinants in K. pneumoniae clinical isolates from Ardabil, Iran. K. pneumoniae is a major cause of infections, including urinary tract infections (UTIs), pneumonia, sepsis, and bloodstream infections. Ciprofloxacin is commonly used as an effective antibiotic for treating recurrent infections caused by this pathogen (7). As a leading cause of healthcare-associated infections worldwide, MDR K. pneumoniae presents significant treatment challenges due to its resistance to antibiotics, chemical disinfectants, and human defense mechanisms such as phagocytosis (8).
Infections caused by K. pneumoniae are associated with high morbidity and mortality rates. Recent epidemiological studies have identified specific genetic characteristics in K. pneumoniae strains linked to hypervirulence. Additionally, Klebsiella species are known to harbor a vast array of antibiotic resistance genes and have played a critical role in the spread of resistance to other gram-negative bacteria. Many of the antibiotic resistance genes now commonly found in multidrug-resistant organisms were first identified in Klebsiella (9).
2. Objectives
Therefore, this study aimed to evaluate the presence of PMQR genes (qnrA, qnrB, qnrC, qnrD, qnrS, oqxA, and oqxB) in clinical samples of K. pneumoniae in Ardabil.
3. Methods
3.1. Bacterial Isolation
From June 2020 to July 2022, 186 K.pneumoniae isolates were collected from clinical specimens of patients admitted to hospitals affiliated with Ardabil University of Medical Sciences, Iran. The isolates were accurately identified using standard microbiological and biochemical tests before being stored in trypticase soy broth (TSB) containing 20% glycerol at -70°C for long-term preservation.
3.2. Ciprofloxacin Susceptibility Tests
The minimum inhibitory concentration (MIC) of ciprofloxacin (Sigma-Aldrich, USA) for K. pneumoniae strains was determined using the standard agar dilution method following the Clinical and Laboratory Standards Institute (CLSI M100-2022) guidelines. Escherichiacoli ATCC 25922 and ATCC 35218 were used as quality control standard strains.
3.3. Detection of Plasmid-Mediated Quinolone Resistance Genes by PCR
The entire plasmid was extracted following the method described by Heringa et al. (10). The quantity and quality of the extracted DNA were assessed using a NanoDropTM 2000/2000c Spectrophotometer (Thermo Fisher Scientific, USA). The extracted DNA was stored at -20°C until further use for gene detection. All collected isolates were screened for the presence of PMQR genes, including qnrA, qnrB, qnrC, qnrD, qnrS, oqxA, and oqxB, using PCR with specific primers (Table 1). The amplification conditions for PCR are detailed in Table 2.
| Genes and Primer Sequence (5′ - 3′) | Annealing (°C) | Product Size (bp) | Ref |
|---|---|---|---|
| qnrA | 58 | 516 | 1 |
| F: ATTTCTCACGCCAGGATTTG | |||
| R: GATCGGCAAAGGTTAGGTCA | |||
| qnrB | 53 | 469 | 1 |
| F: GATCGTGAAAGCCAGAAAG | |||
| R: ACGATGCCTGGTAGTTGTCC | |||
| qnrC | 50 | 447 | 4 |
| F: GGGTTGTACATTTATTGAATC | |||
| R: TCCACTTTACGAGGTTCT | |||
| qnrD | 60 | 636 | 1 |
| F: ATGGAAAAGCACTTTATCAATGA | |||
| R: AACAATAACACCTAAACTCTCAACAA | |||
| qnrS | 60 | 255 | 2 |
| F: TCGGCACCACAACTTTTCAC | |||
| R: TCACACGCACGGAACTCTAT | |||
| oqxA | 64 | 489 | 2 |
| F: CTCTCCTTTCTGCTCGTCGG | |||
| R: AATAGGGGCGGTCACTTTGG | |||
| oqxB | 60 | 240 | 3 |
| F: CGAAGAAAGACCTCCCTACCC | |||
| R: CGCCGCCAATGAGATACA |
Primer Sequences and PCR Program
| Step | Temperatures and Times | Cycles |
|---|---|---|
| Initial denaturation | 4 min at 94°C | 1 |
| Denaturation | 1 min at 94°C | 30 |
| Annealing | 1 min (temperatures are shown for each primer in Table 1) | |
| Extension | 1 min at 72°C | |
| Final extension | 1 min at 72°C | 1 |
The Amplification Program for PCR
The PCR protocol was conducted in a total volume of 25 μL using Ampliqon master mix (Denmark). Each primer was included at a concentration of 10 μmol/L, and 3 μL of extracted DNA was used as the template for amplification. The PCR reaction was performed in a thermal cycler, which amplified the target DNA sequences through different temperature stages according to the specified protocol. Following PCR, the amplified products were separated on a 1% agarose gel in 0.5x TBE buffer and visualized based on fragment size. The PCR products were subsequently confirmed by sequencing to ensure the correct DNA sequence had been amplified.
3.4. Data Analysis
In this study, the researchers collected data on the MICs of ciprofloxacin against K. pneumoniae strains harboring PMQR genes. The data were analyzed using SPSS software version 16. The chi-square test was applied to assess the correlation between the presence of PMQR genes and the MICs of ciprofloxacin. A P-value of < 0.05 was considered statistically significant, indicating a significant association between the presence of PMQR genes and the level of resistance to ciprofloxacin in K. pneumoniae strains.
3.5. Ethics Statement
The study adhered to the ethical principles outlined in the 1975 Declaration of Helsinki and received approval from the Ethics Committee of Ardabil University of Medical Sciences under the ethical code IR.ARUMS.REC.1402.136. Written informed consent was obtained from all participants included in the study.
4. Results
In the present study, among 186 K. pneumoniae clinical isolates, 51.6% (n = 96) were resistant, 11.29% (n = 21) were intermediate, and 37.09% (n = 69) were susceptible to ciprofloxacin. The prevalence of the qnrA, qnrB, qnrS, oqxA, and oqxB genes in K. pneumoniae clinical isolates was 20.9% (n = 39), 49.4% (n = 92), 51.6% (n = 96), 47.8% (n = 89), and 47.3% (n = 88), respectively. The qnrC and qnrD genes were not detected in any of the isolates. Additionally, the prevalence of qnrA, qnrB, qnrS, oqxA, and oqxB genes in ciprofloxacin-resistant K. pneumoniae isolates was 40.6% (n = 39), 80.2% (n = 77), 90.6% (n = 87), 82.3% (n = 79), and 82.3% (n = 79), whereas in the other isolates, the prevalence was 0% (n = 0), 16.6% (n = 15), 10% (n = 9), 11.1% (n = 10), and 10% (n = 9), respectively.
As shown in Table 3, the presence of qnrA, qnrB, qnrS, oqxA, and oqxB genes was significantly associated with increased MIC values and resistance to ciprofloxacin in K.pneumoniae isolates (P < 0.05).
| Gene | Ciprofloxacin (MIC) | |
|---|---|---|
| ≤ 0.5 | 1 ≤ | |
| qnrA+ | 0 | 39 |
| qnrA- | 90 | 57 |
| qnrB+ | 15 | 77 |
| qnrB- | 75 | 19 |
| qnrS+ | 9 | 87 |
| qnrS- | 81 | 9 |
| oqxA+ | 10 | 79 |
| oqxA- | 80 | 17 |
| oqxB+ | 9 | 79 |
| oqxB- | 81 | 17 |
Association Between Ciprofloxacin Resistance and Plasmid-Mediated Quinolone Resistance Genes in Clinical Isolates of Klebsiella pneumoniaa
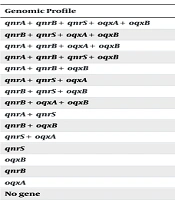
The MIC range for ciprofloxacin was 0.125 - 16 μg/mL. The analysis of ciprofloxacin-resistant genes in K.pneumoniae isolates resistant to ciprofloxacin revealed 15 gene patterns. An increase in the MIC level of ciprofloxacin was observed in isolates harboring multiple PMQR genes. The most frequent genomic profiles were qnrA+ qnrB+ qnrS+ oqxA+ oqxB and qnrB+ qnrS+ oqxA+ oqxB. Isolates that simultaneously carried the qnrA, qnrS, and oqxA genes exhibited the highest MIC values against ciprofloxacin (Table 4).
| Genomic Profile | N = 186 | MIC90 |
|---|---|---|
| qnrA+ qnrB+ qnrS+ oqxA+ oqxB | 31 (16.66) | 16 |
| qnrB+ qnrS+ oqxA+ oqxB | 27 (14.51) | 2 |
| qnrA+ qnrB+ oqxA+ oqxB | 2 (1.07) | 8 |
| qnrA+ qnrB+ qnrS+ oqxB | 1 (0.53) | 2 |
| qnrA+ qnrB+ oqxB | 2 (1.07) | 4 |
| qnrA+ qnrS+ oqxA | 2 (1.07) | 16 |
| qnrB+ qnrS+ oqxB | 9 (4.83) | 4 |
| qnrB+ oqxA+ oqxB | 4 (2.15) | 4 |
| qnrA+ qnrS | 2 (1.07) | 2 |
| qnrB+ oqxB | 5 (2.68) | 4 |
| qnrS+ oqxA | 13 (6.98) | 4 |
| qnrS | 11 (5.91) | 0.5 |
| oqxB | 6 (3.22) | 0.5 |
| qnrB | 11 (5.91) | 0.5 |
| oqxA | 8 (4.3) | 0.5 |
| No gene | 52 (27.95) | 0.25 |
Association Between Ciprofloxacin MIC90 and Genomic Profile of Klebsiella pneumoniae Isolates a
5. Discussion
In recent years, numerous cases of pathogenic drug-resistant bacteria have emerged due to the indiscriminate and arbitrary use of antibiotics. This has led to treatment failures, increased complications, and high healthcare costs. In this context, resistance to quinolone antibiotics, including ciprofloxacin, has risen significantly over the years (11). Resistance to ciprofloxacin in K. pneumoniae infections, a common cause of hospital-acquired infections, poses serious clinical implications, including limited treatment options (12). As a result, the growing prevalence of ciprofloxacin-resistant infections is particularly concerning, especially during antibiotic therapy in hospitalized patients. Resistant strains often require more complex and costly treatment regimens, leading to prolonged hospital stays and increased healthcare expenses (13). Furthermore, these resistant strains can spread within healthcare settings, potentially causing outbreaks in at-risk populations. Patients with ciprofloxacin-resistant K. pneumoniae infections may experience increased morbidity and mortality, as they often present with more severe clinical conditions and require intensive treatment (14). Overall, resistance patterns to ciprofloxacin in K. pneumoniae significantly impact treatment strategies and clinical outcomes, emphasizing the need for careful monitoring and reassessment of treatment protocols to combat antibiotic resistance effectively.
In the present study, the resistance rate to ciprofloxacin among K. pneumoniae clinical isolates was found to be 51.6%. In comparison, Yuan et al. reported a resistance rate of 66% to ciprofloxacin (15), while Saadatian et al. observed a resistance rate of 68.7% in K. pneumoniae isolates (16), which aligns with our findings. Variations in geographical regions may contribute to slight differences in resistance rates.
Studies indicate that qnr genes play a crucial role in the development of resistance to quinolones, including ciprofloxacin. In this study, the prevalence rates of qnrA, qnrB, and qnrS genes in K.pneumoniae clinical isolates were 20.9%, 49.4%, and 51.6%, respectively. According to our findings, a study conducted by Abosadegh et al. in Tehran in 2019 reported the qnrS gene as the most prevalent (35%), followed by qnrB (31%) and qnrA (13%) (17). Similarly, a study conducted in Iraq in 2020 identified qnrS and qnrB genes in 76% and 36% of isolates, respectively, while qnrA, qnrC, and qnrD genes were absent in all isolates (18). Differences in the prevalence of resistance genes across various regions in Iran and worldwide are likely influenced by factors such as geographical variations, sample types, antibiotic usage patterns, accessibility to broad-spectrum and newer antibiotics, genetic variations in bacterial strains, and disparities in antibiotic prescribing practices based on regional healthcare policies.
The oqxAB efflux pumps, associated with the oqxA and oqxB genes and located on the PolA52 plasmid, are among the primary factors contributing to antibiotic resistance in bacteria, particularly resistance to ciprofloxacin (19). In the present study, the prevalence rates of the oqxA and oqxB genes were 47.8% and 47.3%, respectively, which are somewhat lower than those reported in a study by Zomorrodi et al., where the oqxA and oqxB genes were detected in 69.7% and 72.1% of isolates, respectively (20). Rodriguez-Martinez et al. identified concurrent signals of oqxA and oqxB in both chromosomal locations and large plasmids (21). Other studies have reported oqxAB gene prevalence rates ranging from 74% to 100%. Thus, the approximate 47% prevalence of oqxAB genes identified in this study, similar to the findings of Yang et al., is relatively low (1). This discrepancy may be attributed to the fact that, in this study, as in the study by Yang et al., only plasmid DNA was purified, whereas other studies analyzed the whole genome to identify the oqxAB gene. Additionally, this difference may indicate variations in the prevalence of efflux pump-encoding genes among hospital strains due to epidemiological factors (1).
It is important to acknowledge the limitations of this study. Conducted in a single hospital, the isolates examined may not fully represent the broader clinical population, potentially limiting the generalizability of the findings. Furthermore, the study did not analyze the clonal relationships among PMQR-positive isolates. The co-location of the qnr gene with other PMQR genes also requires confirmation through PCR or Southern blot hybridization using specific DNA probes from a single plasmid.
To validate these results, nationwide epidemiological surveys and additional molecular studies are necessary to investigate the potential horizontal transfer of PMQR genes. Other limitations include insufficient coordination among hospitals for sample collection and the relatively small sample size from Ardabil, Iran. Future studies should aim to increase the sample size to enhance the robustness and generalizability of the findings.
5.1. Conclusions
Understanding the frequency of qnr genes and efflux pumps in clinical samples is crucial for selecting appropriate treatment regimens and mitigating the increasing trend of antibiotic resistance. Consequently, the findings of this study can assist healthcare providers in making informed treatment decisions and in preventing the inappropriate prescription of antibiotics.