1. Background
In intensive care unit (ICU) patients across various countries, the incidence of bloodstream infections caused by microbes poses a significant threat to their health. The susceptibility of these individuals to such infections emphasizes the importance of implementing rigorous infection control measures in healthcare settings worldwide. Previous research has indicated that the occurrence of bloodstream infections in the ICU is higher compared to non-ICU wards (1-3). Compared to other patients, individuals admitted to the ICU are at heightened vulnerability due to their prolonged hospitalization and increased need for intensive care. Understanding the diverse microbial pathogens responsible for these bloodstream infections is crucial for implementing targeted treatment strategies and preventing further complications.
This global issue underscores the need for continuous surveillance, prompt diagnosis, and effective management protocols to safeguard the well-being of ICU patients and mitigate the impact of microbial infections on their recovery outcomes (4). In the high-stakes environment of the ICU, timely and accurate microbial infection and blood culture testing play a crucial role in patient management. Rapid identification of pathogens is essential for guiding targeted antibiotic therapy, which helps prevent the progression of infections and minimizes the potential for antimicrobial resistance. By detecting bloodstream infections early, healthcare providers can intervene promptly, potentially saving lives. Additionally, monitoring trends in microbial patterns through culture testing helps hospitals implement effective infection control measures to prevent outbreaks and protect vulnerable patients. In the fast-paced setting of ICUs, the insights provided by these tests are invaluable in ensuring optimal patient outcomes and minimizing the spread of infections within healthcare facilities (5, 6).
Additionally, during the second rise of the COVID-19 pandemic, there was a noted increase in bloodstream infections and blood culture contaminations in the ICU, particularly in months when the ICU was heavily burdened with COVID-19 patients (7, 8).
2. Objectives
Thus, the purpose of this study was to evaluate the antimicrobial trends in blood cultures in the ICUs (from 2017 to 2021) of the largest hospital in northeastern Iran.
3. Methods
3.1. Study Design and Setting
A retrospective cross-sectional study was conducted using medical records of patients admitted to the ICU of Imam Reza Hospital, Mashhad, Iran, between 2017 and 2021. The study was approved by the Ethics Committee of Mashhad University of Medical Sciences (ethical code: IR.MUMS.REC.1401.262).
3.2. Inclusion and Exclusion Criteria
Patients were included in the study if they met the following criteria:
(1) Developed signs and symptoms of infection 48 to 72 hours after ICU admission.
(2) Had no signs or symptoms of infection at the time of admission, and a change of pathogen was observed in two cultures from the same source.
(3) If transferred from other hospital departments, no infection was recorded during the prior hospitalization period.
Exclusion criteria included: (1) Patients with positive blood cultures who had been hospitalized in the last two months or had been admitted for less than five days; (2) patients who had received antibiotics for possible infectious diseases before ICU admission; (3) patients with incomplete or inconsistent medical records.
3.3. Data Collection
Data were extracted from both paper-based patient records and the hospital information system (HIS). Information collected included: (1) Demographic data: Age, sex; (2) clinical characteristics: Underlying conditions, length of ICU stay; (3) microbiological findings: Culture results, antibiograms.
3.4. Laboratory Investigations
Blood cultures were processed according to hospital laboratory protocols. Pathogen identification and antimicrobial susceptibility testing were conducted using standard microbiological methods, including automated systems and manual techniques when necessary:
- Blood samples were inoculated into blood culture bottles and incubated at 35 ± 2°C in an automated blood culture system.
- Positive cultures were sub-cultured on appropriate agar media for bacterial and fungal isolation.
- Bacterial identification and antimicrobial susceptibility testing were performed using the VITEK 2 system (bioMérieux, France).
- Fungal identification was conducted using microscopic morphology and biochemical assays.
3.5. Statistical Analysis
Data analysis was performed using GraphPad Prism 6.0 and SPSS version 16. Descriptive statistics were used to summarize categorical variables as frequencies and percentages, while continuous variables were expressed as means and standard deviations, where appropriate.
4. Results
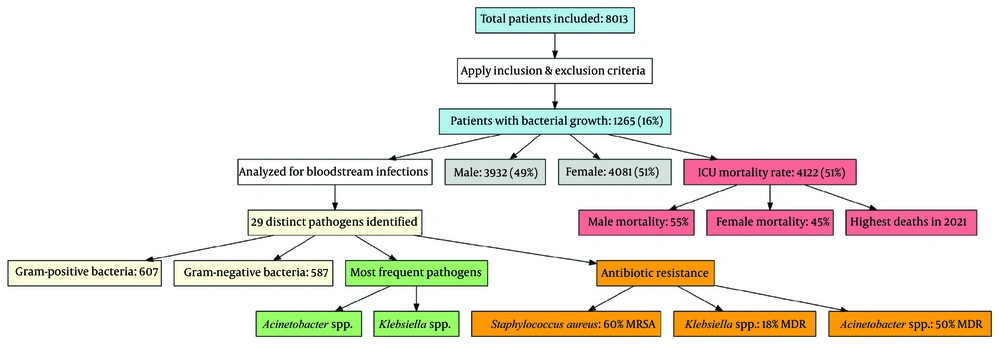
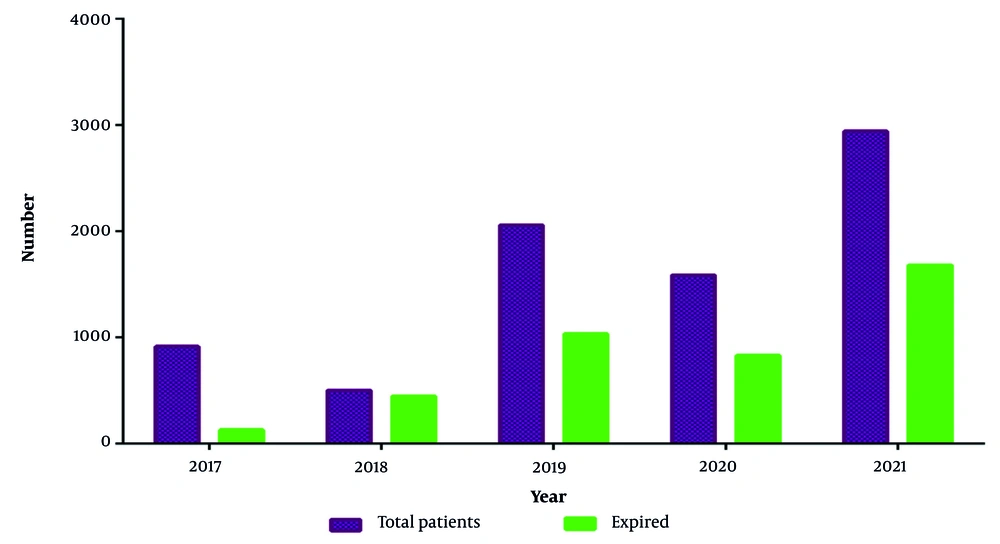
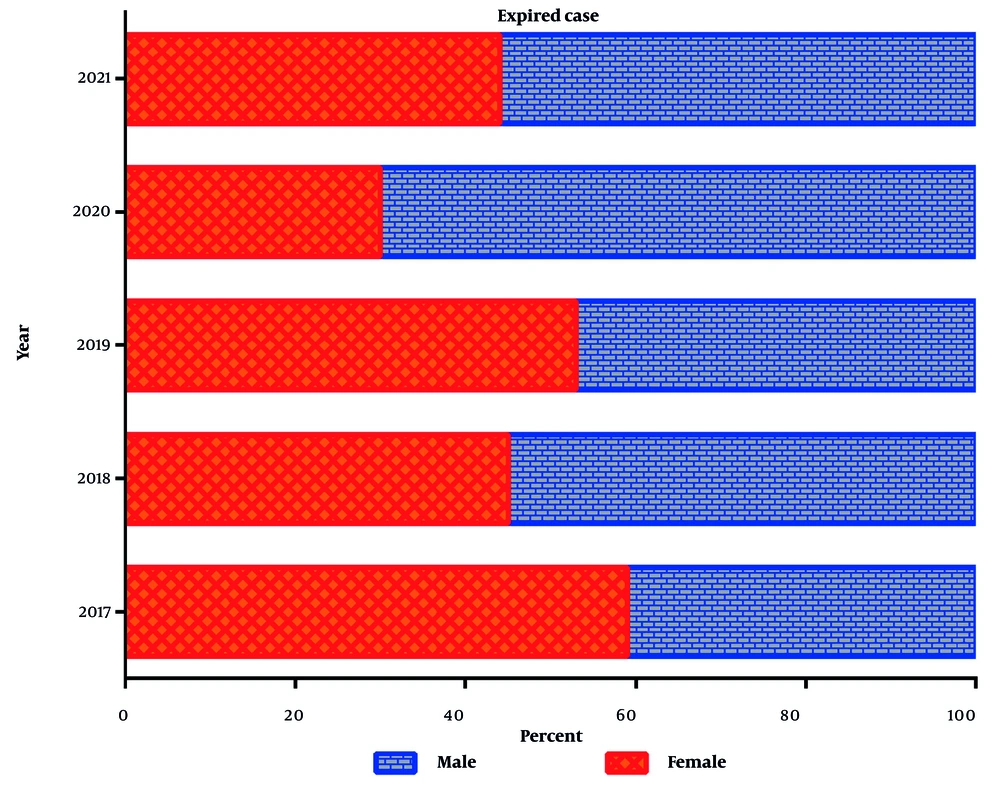
A total of 8013 patients were initially included in the study. After applying the inclusion and exclusion criteria, 1265 patients (16%) exhibited bacterial growth and were analyzed for bloodstream infections. To provide a clearer overview of the study process, a flow diagram (Figure 1) illustrates patient selection, exclusions, and final sample size. Of the total study participants (n = 8013), 3932 (49%) were male. The overall mortality rate in the ICU ward was 51% (n = 4122), with a higher mortality rate among males compared to females (55% vs. 45%) (Figures 2 and 3). The highest number of deaths among ICU patients was recorded in 2021.
Flowchart illustrating the study process, including patient selection, exclusion criteria, bloodstream infection analysis, gender distribution, intensive care unit (ICU) mortality rates, pathogen identification, and antibiotic resistance patterns. The diagram highlights key findings, such as the prevalence of gram-positive and gram-negative bacteria, the most frequently isolated pathogens, and the proportion of multidrug-resistant infections
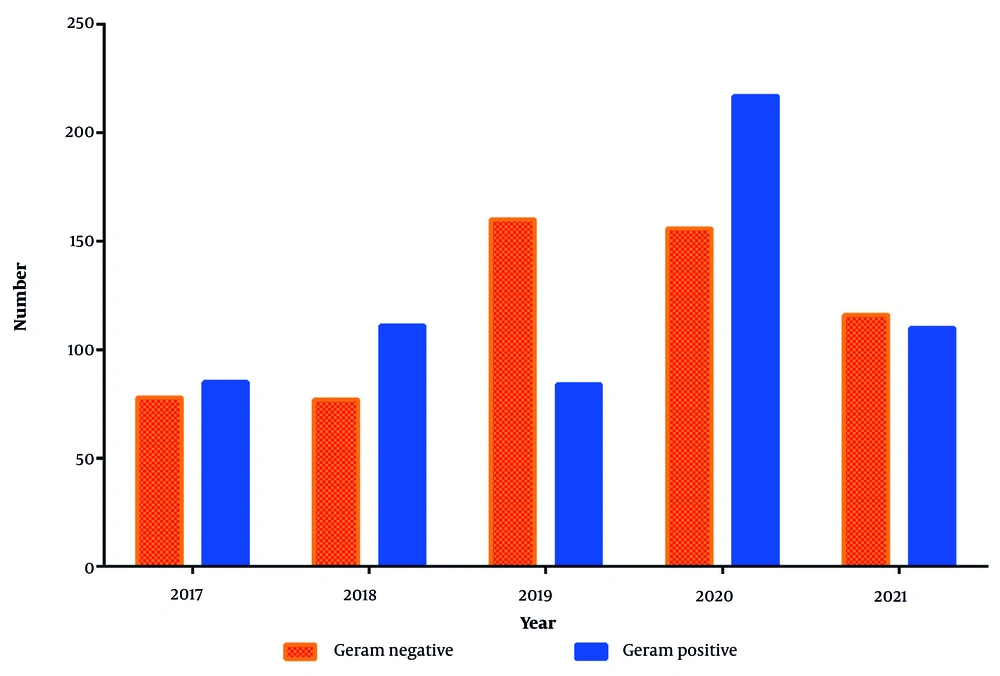
Among the 1265 patients with bloodstream infections, 29 distinct types of pathogens were identified. Gram-positive bacteria (n = 607) were slightly more prevalent than gram-negative bacteria (n = 587) (Figure 4). The most frequently isolated pathogens responsible for bloodstream infections were Acinetobacter spp. and Klebsiella spp., as detailed in Table 1. Notably, 60% of Staphylococcus aureus infections were methicillin-resistant (MRSA). Additionally, 18% of Klebsiella spp. infections and 50% of Acinetobacter spp. infections were classified as multidrug-resistant (MDR) gram-negative bacteria.
| Bacteria | Number of Infected Cases | |||||
|---|---|---|---|---|---|---|
| 2017 | 2018 | 2019 | 2020 | 2021 | 2022 | |
| Acinetobacter spp. | 8 | 8 | 29 | 35 | 35 | 115 |
| Acinetobacter spp. (MDR) | 31 | 32 | 50 | 1 | - | 114 |
| Candida spp. | 7 | 6 | 18 | 43 | 24 | 98 |
| Coagolasenegative staphylococci | 2 | 2 | 1 | 4 | 2 | 11 |
| Corynebacterium spp. | 5 | 3 | 1 | 8 | 11 | 28 |
| Enterobacteraerogenes | 2 | 2 | 6 | 1 | - | 11 |
| Enterococcus spp. | 7 | 14 | 3 | 39 | 26 | 89 |
| Enterococcus spp. (VRE) | 10 | 3 | 5 | 6 | - | 24 |
| Entrobacterspp. (ESBL) | 1 | 1 | 19 | 27 | - | 48 |
| Escherichia coli | 2 | 2 | 2 | 9 | 12 | 27 |
| E. coli (ESBL) | 7 | 6 | 6 | - | - | 19 |
| Klebsiellapneumoniae | 9 | 13 | 3 | 69 | 37 | 131 |
| K. pneumoniae(MDR) | 7 | 4 | 13 | - | - | 24 |
| K.pneumoniae(ESBL) | 2 | 1 | 5 | - | - | 8 |
| Micrococcus spp. | 1 | 18 | 22 | - | 1 | 42 |
| Non fermentergram negative bacilli | 2 | 1 | 1 | - | - | 4 |
| Pseudomonasespp. | 2 | 2 | 13 | - | - | 17 |
| Pseudomonaseaeroginosa | 3 | 3 | 2 | - | - | 8 |
| Staphylococcus epidermidis | 6 | 8 | 2 | 9 | 13 | 38 |
| S. epidermidis (MRS) | 8 | 17 | 5 | 57 | - | 87 |
| S. aureus | 3 | 4 | 6 | - | 32 | 45 |
| S. aureus (MRSA) | 8 | 8 | 25 | 18 | 8 | 67 |
| S.haemolyticus | 14 | 14 | 2 | 57 | 12 | 99 |
| S.saprophyticus | 5 | 1 | 3 | 19 | 2 | 30 |
| S.simulans(MRS) | 2 | 4 | 4 | 2 | 2 | 14 |
| Stenotrophomonas maltophilia | 2 | 2 | 2 | - | 19 | 25 |
| Streptococouslike organism suspected to leuconostocSpp isolated | 1 | 1 | 1 | 5 | - | 8 |
5. Discussion
This study provides a primary analysis of data obtained from the ICUs of a major educational hospital in northeastern Iran. In blood cultures collected from ICU patients before the coronavirus outbreak, a variety of microbial patterns were observed, with common pathogens such as Acinetobacter spp. and Klebsiella spp. being prevalent. However, post-coronavirus, there was a noticeable shift in the microbial landscape. The presence of opportunistic infections like Candida species increased significantly, possibly due to changes in patient immune responses or antibiotic usage. Additionally, a rise in multidrug-resistant organisms was noted, indicating that healthcare-associated infections are becoming more challenging to treat. This shift highlights the complex interplay between infectious agents and external factors like pandemics, emphasizing the need for vigilant surveillance and tailored treatment strategies in critical care settings.
The research conducted from 2017 to 2021 at Imam Reza Hospital in Mashhad, Iran, analyzed microbial patterns in blood cultures collected from ICU patients. The findings revealed a concerning increase in multidrug-resistant strains among the detected microbes, posing challenges for effective treatment strategies. Gram-negative bacteria were predominant, with Acinetobacter spp. showing high prevalence rates in ICU patients, highlighting the escalating threat of antimicrobial resistance in hospital settings. Additionally, fungal species such as Candida albicans were identified, underscoring the importance of antifungal stewardship in managing bloodstream infections. The investigation indicates that targeted infection control methods and antimicrobial treatments are required to tackle the heterogeneous microbial environment identified in ICU patients in Iran.
Particularly in an ICU, the identification and treatment of infections depend heavily on the microbial patterns seen in blood cultures. In order to administer proper and timely therapy to patients with serious illnesses in the busy and demanding ICU of Imam Reza Hospital in Mashhad, Iran, it is essential to fully understand the microbial patterns present in blood cultures (9). Blood culture is a diagnostic test that involves incubating a blood sample in a culture medium to detect the presence of microorganisms, such as bacteria, fungi, or viruses, that may be causing an infection. In the ICU, where patients are often immunocompromised and at a higher risk of developing infections, blood cultures are routinely performed to help identify the causative agent and guide the selection of appropriate antibiotics or antifungal therapy (10).
The microbial patterns observed in blood cultures in the ICU of Imam Reza Hospital can vary depending on various factors, including the patient population, underlying medical conditions, and local epidemiology of pathogens. Gram-positive bacteria like Staphylococcus epidermidis (MRS) and gram-negative bacteria like Acinetobacter spp. and Klebsiella spp. were frequently recovered from blood cultures in the ICU. Fungal pathogens, such as Candida species, were also identified in blood cultures, particularly in patients with prolonged hospital stays or prior exposure to broad-spectrum antibiotics.
According to a study conducted in Gondar, Northwest Ethiopia, the three most frequent bacterial infections responsible for neonatal sepsis were Klebsiella pneumoniae (15.8%), coagulase-negative staphylococci (21.6%), and Staphylococcus aureus (40.8%) (11). These bacteria are known to be common causes of neonatal sepsis worldwide and are often associated with serious complications and poor outcomes. In contrast, among individuals hospitalized in the ICU in Qazvin, Iran, Stenotrophomonas (41.0%) was the most frequently isolated bacterium from bloodstream infections (12). In a 4-year retrospective study conducted in Isfahan, the most common bacteria causing bloodstream infections in patients in both ICU and non-ICU wards were Klebsiella spp., S. epidermidis, and Acinetobacter spp. (13).
In the current study, MRSA accounted for 60% of all S. aureus infections. This percentage is greater compared to rates reported in Canada (22.3%) (14), Europe (39%) (15), and the USA (55%) (16). However, the MRSA percentage in the present investigation is smaller than that of two independent findings from Iran (87.5% - 96.2%) (17, 18).
In the present study, 51% of ICU patients died. According to the findings of Hattori et al., among a sample of 2,105 patients diagnosed with bloodstream infections, a total of 319 individuals passed away, with a 30-day mortality rate reported at 15.2% (19). In Italy, the mortality rates for patients with bloodstream infections were reported as follows: 12% at 7 days, 25% at 30 days, and 36% at 90 days (20). In Ireland, the mortality rate for bloodstream infections was reported to be 15% (21).
The identification of microbial patterns in blood cultures is essential for guiding empiric antibiotic therapy and de-escalating treatment once the causative agent is identified. Timely and appropriate antibiotic therapy is crucial in the ICU setting to prevent the progression of infections, reduce mortality rates, and minimize the development of antibiotic resistance. Therefore, the accurate and prompt identification of microbial patterns in blood cultures is vital for improving patient outcomes and optimizing the administration of antimicrobial medication.
Apart from determining the causative pathogens, blood culture results provide valuable information on antimicrobial susceptibility patterns, which can help guide antibiotic selection and dosing. Antimicrobial resistance is a growing concern in healthcare settings, including ICUs, and the surveillance of resistance patterns in blood cultures can help inform antibiotic stewardship programs and promote the careful application of antibiotics to combat the advancement of multidrug-resistant pathogens.
In conclusion, the microbial patterns observed in blood cultures in the ICU of Imam Reza Hospital in Mashhad, Iran, are essential for the diagnosis and management of infections in seriously ill individuals. Understanding the microbial etiology of bloodstream infections and accurately interpreting blood culture results are crucial for providing optimal patient care, reducing morbidity and mortality rates, and combating antimicrobial resistance. Continued surveillance of microbial patterns in blood cultures and the implementation of antimicrobial stewardship programs are essential components of infection control strategies in the ICU setting. By staying vigilant and proactive in monitoring and responding to microbial patterns in blood cultures, healthcare providers can improve patient outcomes and promote the judicious use of antibiotics in the ICU.