1. Background
The genus Nocardia includes gram - positive, partially acid - fast, strictly aerobic, and filamentous bacteria of the family Nocardiaceae in the order of Actinomycetales (1-5). Members of the genus Nocardia comprise several species that can be found worldwide as saprophytic soil microorganisms (1, 2, 5). Nocardia species are the causes of a variety of infections in humans that include cutaneous, pulmonary, ocular, cerebral, and systemic nocardiosis (5) and human is infected through aerosols or scratch (5). Since the growth of Nocardia is very slow and it may be covered by normal microflora and other rapidly growing bacteria, the isolation can be difficult (6). Antibacterial therapy of Nocardia infections depends on the organ or site and correct diagnosis of Nocardia species (7, 8). However, regarding the numerous reports about Nocardia in Iran, rapid and accurate identification of Nocardia spp. is of importance. One hundred twenty - seven clinical isolates of Nocardia from different major cities of Iran were studied between 2009 and 2015. Nocardia asteroides was the most frequently recovered, followed by Nocardia farcinica and Nocardia cyriacigeorgica (9). Other reports have shown N. cyriacigeorgica, N. asteroides complex, Nocardia nova complex, Nocardia otitidiscaviarum, and Nocardia transvalensis in Iran (10-13). The prevalence of Nocardial infections in another study was 5.28% (6). Recognition of Nocardia to species level is important for the four reasons: definitive and accurate identification, predicting antimicrobial susceptibility, epidemiological purposes, and effective treatment (14-16). The identification of Nocardia used to be conducted via traditional methods (colonial and microscopic morphology and biochemical tests) (15, 17). The routine identification of Nocardia species is difficult (18). Due to time - consuming bacterial culture and insufficient phenotypic, the use of rapid, sensitive, and accurate molecular methods for accurate diagnosis of infections and bacterial species seems necessary (5, 14). Today, molecular techniques developed for accurate identification include PCR sequencing alone or with other molecular analyses such as PRA (PCR restriction enzyme pattern analysis) that is one of the first molecular techniques for the identification of Nocardia species (1, 19). The RFLP method is a PCR reaction based on restriction enzyme digestion. The use of molecular approaches such as PCR - RFLP has been the focus of recent investigations to distinguish the newer species of Nocardia isolates from other actinomycetes genera or Mycobacteria species (14, 20). In the current study, hsp65 gene was used for the identification purpose by the RFLP technique. There are molecular methods relying on sequencing of one of the most important genes to determine the species. The 16S rRNA gene is valuable for the identification and taxonomic classification because this gene is a housekeeping and conserved gene that is in all bacteria. The fragment of 16S rRNA gene (1500bp) is a conserved and variable area that is used for taxonomic comparisons. The use of 16S rRNA gene sequencing in the clinical laboratory is beneficial for identifying unknown bacteria (21). In the current study, the polymerase chain reaction - restriction fragment length polymorphism (PCR - RFLP) of hsp65 gene and sequencing of 16S rRNA gene analyses were performed for a definitive identification of Nocardia species of soil.
2. Methods
According to our previous study, soil samples were randomly collected from 4 cm deep from various geographical regions of Iran. The samples in sterile plates were transferred to actinomycetes lab within 24 - 48 hours. Strains of Nocardia were isolated by paraffin baiting technique. All isolates were identified to the species level by biochemical tests in our previous study (17, 22). Then, PCR - RFLP of the hsp65 gene and sequencing of 16S rRNA gene were done for accurate identification of Nocardia isolates.
2.1. DNA Extraction
DNA extraction was done by boiling in STET solution. STET buffer contained Tris - HCl (10 mM), NaCl (0.1 M), EDTA (1 Mm), and Triton X100 (%5 (v/v)) for 100 ml STET “pH = 8”. Briefly, the isolates of Nocardia were cultured on nutrient agar medium and incubated at 35°C for 5 days. A loop full of the pure colony of bacteria was suspended in 200 µl (microliters) of STET buffer and boiled at 100°C for 30 min and centrifuged at 10,000 rpm for 10 min. The supernatant was transferred into a sterile micro tube, added by 500 ml cold ethanol, and incubated for 60 min at -20°C. After this stage, the microtube was centrifuged for 10 min at 13,000 rpm and the supernatant was discarded. DNA pellets were dried and dissolved in 50µl (microliters) sterile distilled water and stored at -20°C (23).
2.2. PCR - RFLP (Polymerase chain reaction - restriction fragment length polymorphism)
2.2.1. Amplification of a Portion of the hsp Gene
After DNA extraction, a 440 - bp fragment of the hsp65 gene encoding the 65 - kDa heat shock protein from Nocardia strains was amplified by using two primers described by Telenti et al. (24). The following primers were used: a forward primer with the sequence 5' - ACCAACGATGGTGTG TCCAT - 3' (TB11); a reverse primer with the sequence 5' -CTTGTCGAACCG CATACCCT - 3' (TB12) (24). Reaction mixtures consisted of 2.5 U of Taq polymerase, 1.5 mM MgCl2 (50mM), 0.3mM each primer, 0.2 mM each deoxynucleoside, 1 × PCR buffer and 5ml of DNA extract. The PCR amplification was performed in a total volume of 50 ml. The temperature cycling for amplification was performed in the initial denaturation step (94°C for 5 min), 40 cycles of amplification (94°C, 55°C, and 75°C for 1 min at each temperature), and the final extension step (72°C for 10 min). The amplicons were electrophoresed on 1.5% agarose gel with ethidium bromide. We considered H37RV strain of Mycobacterium tuberculosis (manufacturer of the 441bp fragment) as a positive control at PCR reaction.
2.3. Restriction Fragment Length Polymorphism (RFLP)
After amplification, the PCR products were digested with HinfI (Biolabs Co., England), BstEII, and MspI (Fermentas Co.) restriction enzymes separately (14, 18). Enzymatic digestion of PCR products was performed under appropriate conditions (according to the enzyme manufacturer instructions). All the three digest enzymes showed the highest activity at 37 °C. RFLP products were electrophoresed in 3% agarose gel (Invitrogen, USA) in 0.5 × TBE buffer (Tris - Borate - EDTA) at 50 V for 6 h, followed by staining with ethidium bromide. For determining the size of the fragments generated by PCR - RFLP, we used a 50bp molecular marker (Geneon - Germany).
2.4. Amplification of a Portion of the 16S rRNA Gene
The 1500 - bp fragment of the 16S rRNA gene was amplified using two universal primers including a primer with the sequence 5' - AGAGTTTGATCMTGGCTCAG - 3' (27F) and a primer with the sequence 5' - AAGGAGGTGWTCCARCC - 3' (1525R) (2). Reaction mixtures consisted of 2.5 U of Taq polymerase, 1.5 mM MgCl2 (50mM), 0.3mM each primer, 0.2 mM each deoxynucleoside, 1 × PCR buffer, and 5ml of extracted DNA. The temperature cycling for amplification was performed in the initial denaturation step (94°C for 5 min), 32 cycles of amplification (94°C for 5 min; 55°C for 1 min, and 75°C for 90 sec), and the final extension step (72°C for 10 min). PCR products were electrophoresed in 1.5% agarose gel with ethidium bromide.
2.5. Sequencing of the 1500 - Bp Fragment of the 16S rRNA Gene
The agarose gel containing 16S rRNA gene of band 1500 bp was cut and sent to Bioneer company (Korea) for sequencing.
2.6. Sequence Analysis of 16S rRNA Gene
The 16S rRNA gene sequences of Nocardia isolates were aligned with the sequences of Nocardia species (retrieved from GenBankTM database) using the jPhydit software according to instructions. The sequences of strains were analyzed through BLAST search whit the sequences of bacteria available in databanks of NCBI (25). The phylogenetic tree was plotted using MEGA 5 software (26).
3. Results
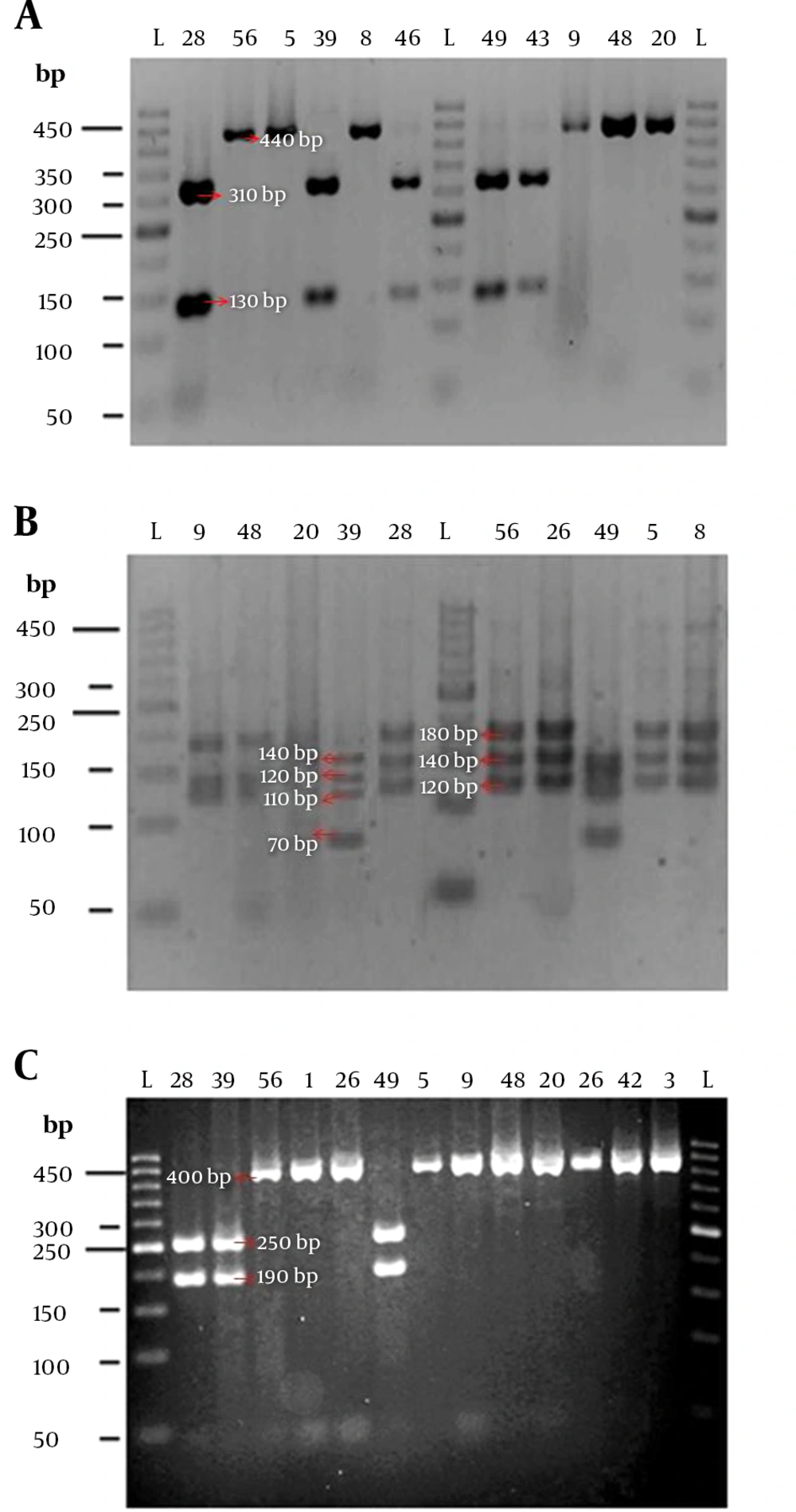
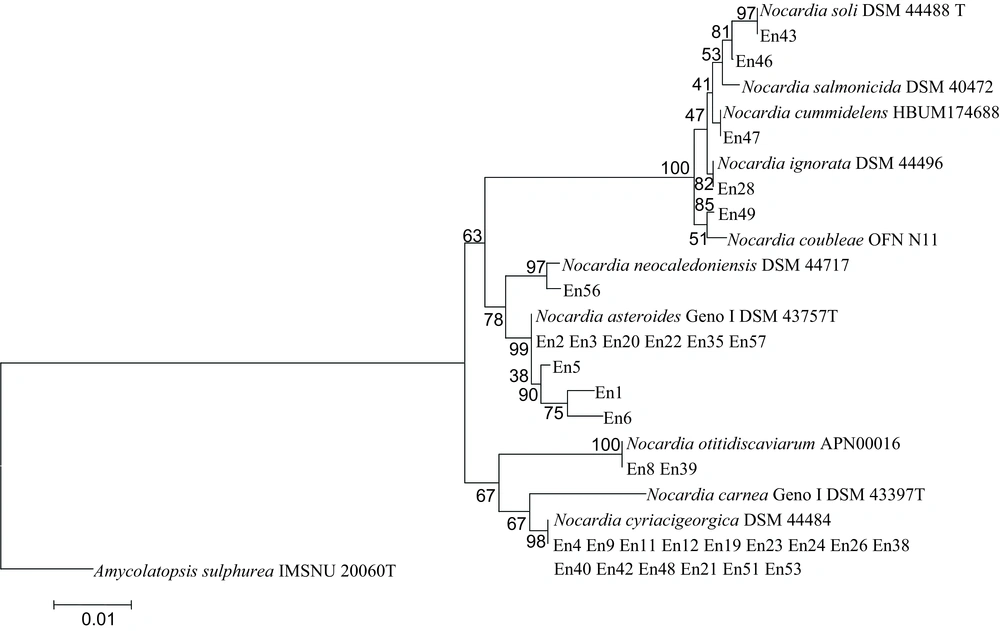
We present an analysis of 32 Nocardia isolates from soil in Iran using banding patterns produced by single digestions of 440 - nt fragments of the hsp65 gene with endonucleases BstEII, MspI, and HinfI. The restriction digest is then separated and visualized by agarose gel electrophoresis (Figure 1). After PCR - RFLP analysis of the hsp65 gene, four different RFLP profiles were revealed based on the fragments sizes, which were numbered from A to D. The restriction profiles of each isolate for each used enzyme are shown in Table 1. The RFLP profiles obtained in this study were compared with the previously known patterns by Rodriguez et al. (14). Thus, the profile BstEII, 440; MspI, 120/140/180; and HinfI, 440 obtained in this study (pattern C in the table) determined a group of Nocardia spp. including Nocardia abscessus, Nocardia araoensis, Nocardia asiatica, Nocardia asteroides type IV, N. asteroides type IVATCC 14759, Nocardia brevicatena, N. cyriacigeorgica, Nocardia higoensis, Nocardia neocaledoniensis, Nocardia paucivorans, Nocardia puris, and Nocardia vinacea according to Rodriguez et al. (14). Since some of the Nocardia species have similar PCR-RFLP patterns, we performed PCR - RFLP analysis to reveal biochemical and morphological characteristics (17). After RFLP - PCR analysis, 9 isolates (En1, En2, En3, En5, En6, En20, En22, En35, and En57) as N. asteroides drug pattern type VI, and 14 isolates (En4, En9, En11, En19, En21, En23, En24, En26, En38, En40, En42, En48, En51, and En53) as N. cyriacigeorgica, and 1 isolate (En8) as N. otitidiscaviarum were identified. The results of PCR - RFLP of hsp65 gene in all isolates were similar to the results of phenotypic tests and sequencing of 16S rRNA gene. The results of RFLP and sequence analysis of these isolates are presented in Table 2. Isolate En56 by sequence analysis of 16S rRNA was determined as N. neocaledoniensis while, based on its hsp65 - RFLP pattern, it was identified as N. asteroides (14). In some strains, 440 bp fragment was not digested by BstEII enzyme, whereas others were digested into two fragments with 130bp and 310bp size range (Figure 2A); these fragments have not been reported so far. It is noteworthy that after PCR - RFLP of PCR products, only the enzyme BstEII created new fragments. Finally, in these isolates, two PCR - RFLP profiles (new pattern A and B) were detected by a combination of BstEII, MspI, and HinfI digest analysis. A new pattern A (BstEII, 130/310; MspI, 120/140/180; HinfI, 190/250) in isolate 28 and a new pattern B (BstEII, 130/310; MspI, 70/110/120/140; HinfI, 190/250) in isolates En39, En43, En46, En47, and En49 were reported. For accurate identification, these isolates underwent 16S rRNA gene sequencing. These isolates after sequence analysis of 16S rRNA gene were diagnosed as N. otitidiscaviarum, N. soli, Nocardia cummidelens, and N. coubleae (See Table 2). The new restriction patterns were added to the previously published patterns. The phylogenetic tree of the 16S rRNA gene of our isolates was constructed using the neighbor - joining algorithm with bootstrap analysis for 1000 replicates in the MEGA 5 software (Figure 3).
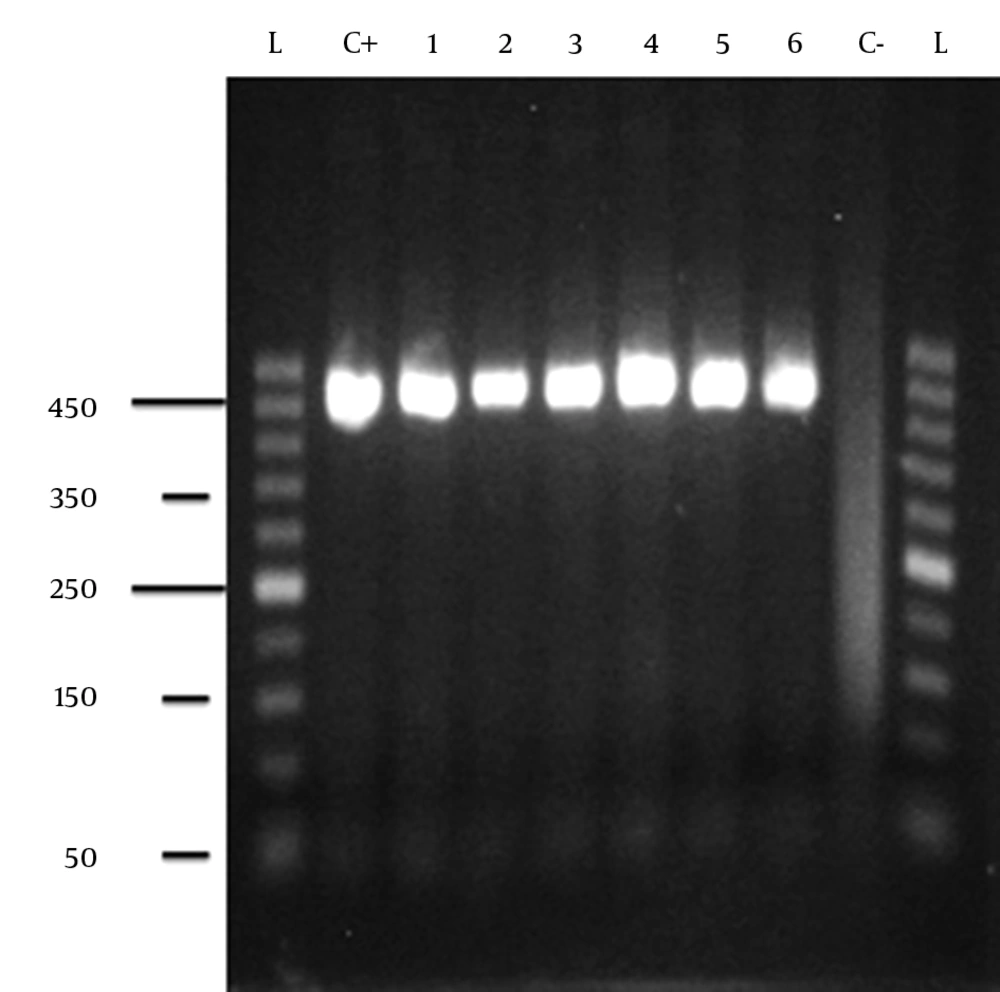
Agarose gel electrophoresis of PCR products amplified from hsp65 gene of Nocardia isolates with Telenti primers. Lanes labeled from left to right: L: generuler 50 bp ladder; C+, positive control (H37Rv); lanes 1 through 6, isolates number; C-,negative control (no DNA). The numbers are molecular sizes in kilobases.
hsp65 RFLP patterns observed with BstEII (A), MspI (B), and HinfI (C) for the randomly selected Nocardia isolates on 3% agarose gels. L, GeneRuler 50 bp ladder; numbers from left to right, isolates number; bp, base pairs. The expected sizes of the products are indicated in Table 1.
| Fragment Sizes (Bp) Obtained by: | Pattern Nameb | Identification of Nocardia Spp. | The En Strains Exhibiting Each of the Four hsp65 Gene Banding Patterns (A - D) | ||
|---|---|---|---|---|---|
| BstEII | MspI | HinfI | |||
| 130/310 | 120/140/180 | 190/250 | Ac | Unkouwn | En28 |
| 70/110/120/140 | 190/250 | Bd | Unkouwn | En39, En43, En46, En47 and En49 | |
| 440 | 120/140/180 | 440 | Ce | N. abscessus, N. araoensis, N. asiatica, N. asteroides type IV, N. asteroides type IVATCC 14759, N. brevicatena, N. cyriacigeorgica, N. higoensis, N. neocaledoniensis, N. paucivorans, N. puris, N. vinacea | En1, En2, En3, En5, En6, En12, En20, En22, En35, En57, En4, En9, En11, En19, En21, En23, En24, En26, En38, En40, En42, En48, En51, En53 and En56 |
| 110/330 | De | N. brasiliensis, N. inohanesis, N. niigatensis, N. otitidiscaviarum, N. shimofusensis, N. yamanashiensis | En8 | ||
aWe identified Nocardia spp. based on the 16S rRNA gene sequencing; please, refer to Table 2 for Genbank accession number on the 16S rRNA gene sequence.
bFour patterns (A to D) were observed for hsp65 gene digested by three restrictions BstEII, MspI, and HinfI with comparison of fragment sizes from restriction in this study.
c“A” and “B” are new RFLP patterns. Therefore, the identification of these isolates to the species level based on this method alone was not possible.
d“A” and “B” are new RFLP patterns. Therefore, the identification of these isolates to the species level based on this method alone was not possible.
eThe RFLP patterns “C’’ and “D” according to the database published by Rodríguez et al. (2006) belonged to 12 species; therefore, the identification of these strains based on RFLP pattern was performed with respect to the phenotypic features (in Table 2).
| Strain(S) (Isolate [No. of Isolates]) | Phenotypic Test Resultsa | RFLP Pattern of hsp65 Geneb | RFLP Results (Species) | Sequencing Results of 16s rRNA(Species Designation of Strain) | Genbank Accession Number on the 16S rRna Gene Sequence |
|---|---|---|---|---|---|
| En28 | N. africana | A | Not identified | N. ignorata | KP137541 |
| En39 | N. otitidiscaviarum | B | Not identified | N. otitidiscaviarum | KP137543 |
| En43 | N. otitidiscaviarum | B | Not identified | N. soli | KP137544 |
| En46 | N. coubleae | B | Not identified | N. cummidelens | KP137545 |
| En47 | N. asteroides | B | Not identified | N. cummidelens | KP137546 |
| En49 | N. asteroides complex | B | Not identified | N. coubleae | KP137547 |
| En56 | N. asteroides | C | N. asteroides | N. neocaledoniensis | KP137548 |
| En12 | N. africana | C | N. cyriacigeorgica | N. cyriacigeorgica | KP137529 |
| En1 | N. asteroides | C | N. asteroides | N. asteroides | KP137517 |
| En2 | N. asteroides | C | N. asteroides | N. asteroides | KP137518 |
| En3 | N. asteroides | C | N. asteroides | N. asteroides | KP137519 |
| En5 | N. asteroides | C | N. asteroides | N. asteroides | KP137524 |
| En6 | N. asteroides | C | N. asteroides | N. asteroides | KP137525 |
| En20 | N. asteroides | C | N. asteroides | N. asteroides | KP137520 |
| En22 | N. asteroides | C | N. asteroides | N. asteroides | KP137521 |
| En35 | N. asteroides | C | N. asteroides | N. asteroides | KP137522 |
| En57 | N. asteroides | C | N. asteroides | N. asteroides | KP137523 |
| En4 | N. cyriacigeorgica | C | N. cyriacigeorgica | N. cyriacigeorgica | KP137526 |
| En9 | N. cyriacigeorgica | C | N. cyriacigeorgica | N. cyriacigeorgica | KP137527 |
| En11 | N. cyriacigeorgica | C | N. cyriacigeorgica | N. cyriacigeorgica | KP137528 |
| En19 | N. cyriacigeorgica | C | N. cyriacigeorgica | N. cyriacigeorgica | KP137530 |
| En21 | N. cyriacigeorgica | C | N. cyriacigeorgica | N. cyriacigeorgica | KP137538 |
| En23 | N. cyriacigeorgica | C | N. cyriacigeorgica | N. cyriacigeorgica | KP137531 |
| En24 | N. cyriacigeorgica | C | N. cyriacigeorgica | N. cyriacigeorgica | KP137532 |
| En26 | N. cyriacigeorgica | C | N. cyriacigeorgica | N. cyriacigeorgica | KP137533 |
| En38 | N. cyriacigeorgica | C | N. cyriacigeorgica | N. cyriacigeorgica | KP137534 |
| En40 | N. cyriacigeorgica | C | N. cyriacigeorgica | N. cyriacigeorgica | KP137535 |
| En42 | N. cyriacigeorgica | C | N. cyriacigeorgica | N. cyriacigeorgica | KP137536 |
| En48 | N. cyriacigeorgica | C | N. cyriacigeorgica | N. cyriacigeorgica | KP137537 |
| En51 | N. cyriacigeorgica | C | N. cyriacigeorgica | N. cyriacigeorgica | KP137539 |
| En53 | N. cyriacigeorgica | C | N. cyriacigeorgica | N. cyriacigeorgica | KP137540 |
| En8 | N. otitidiscaviarum | D | N. otitidiscaviarum | N. otitidiscaviarum | KP137542 |
A 1500 - bp fragment of the 16S rRNA gene sequence - based phylogenetic tree of Nocardia isolates with those of closely related species computed by the Neighbor - joining (NJ) analyses and Kimura 2 - parameter (K2P) model. The support of each branch determined from 1000 bootstrap samples. Bar 0.01 indicates one nucleotide suBstitution per 100 nucleotides.
4. Discussion
The use of molecular methods for identification of Nocardia is based on restriction endonuclease digestion, nucleic acid amplification, or nucleotide sequencing techniques. For accurate diagnosis of Nocardia spp. due to their diversity, we cannot rely on using phenotypic or genotypic methods alone (27). A combination of molecular and phenotypic methods is efficient in detection and identification of Nocardia species. To date, the Nocardia genus comprises 113 species (28). The present study was done to identify Nocardia species based on profiles obtained from PCR - RFLP of the hsp65 gene and sequencing of the 16S rRNA gene. The most commonly used enzymes in Nocardia are MspI, HinfI, BstEII, and BsaHI (14, 18). PCR - RFLP analysis of the groEl gene has been done to distinguish Nocardia isolates from the genus Mycobacterium (29). Steingrube et al. reported 12 species and taxa of Nocardia by a PRA method using the MspI and BsaHI enzymes (30). Thus, the PRA (PCR restriction enzyme pattern analysis) allowed the identification of the drug pattern types in Nocardia spp. In addition, PCR - RFLP of the hsp65 gene can lead to a rapid, presumptive species identification (14). Here, we have earned the pattern profiles of the isolates of Nocardia. In the present study, the results obtained from biochemical tests of the strains of Nocardia from soil (17) were compared with the results obtained from PCR - based RFLP assay. It appears that PCR - based RFLP analysis is a useful tool to detect and distinguish Nocardia spp. from the soil and clinical specimens and distinguish Nocardia from another similar genus such as Mycobacterium, Rhodococcus, Dietzia, etc. (20, 31). In a study, biochemical tests, amplification, and REA (Restriction Endonuclease Analysis) of portions of the 16S rRNA gene (digested by HinP1I and DpnII) and hsp65 gene (digested by MspI, HinfI, and BsaHI) were done to detect 28 isolates of Nocardia and one isolate was not identifiable by its HSP gene RFLP pattern (18). In our study, seven strains (En12, En28, En39, En43, En46, E47, and En49) produced hsp65 restriction-endonuclease fragment patterns that were not in agreement with their identification by biochemical/phenotypic tests (Table 2). RFLP patterns in these strains, except strain En12, were different from other known PCR - RFLP profiles. In this study, 25 isolates showed PCR - RFLP patterns and sequencing of 16S rRNA gene results similar to biochemical tests (Table 2). Significantly, each of these approaches has its advantages and disadvantages to identify species. On the other hand, any of them are usually not sufficient to identify all strains of Nocardia alone. For instance, one of the advantages of the PRA technique (PCR - RFLP analysis targeting hsp65 gene) is differentiation of the genus Mycobacterium of Nocardia species (32). Another study using the hsp65 PCR - RFLP method and restriction enzymes MspI, HinfI, BsaHI, HaeIII, and BstEII identified nine isolates to species and biotype levels. In addition, three isolates were identified to species level and two isolates to genus level (20). Rodriguez - Nava et al. focused on PRA of the hsp65 gene using BstEII, MspI, and HinfI to differentiate Nocardia species and reported the restriction patterns of 36 species of Nocardia many of which had the same restriction patterns (14). Some of Nocardia species have similar biochemical and morphological features and PCR - RFLP profiles. It seems further testing for species detection probabilities such as DNA sequencing is essential especially for new species (18). The 16S rRNA gene sequence plays an important role in species identification and taxonomy. This gene is used for phylogenetic studies (31, 33). By using BstEII digests alone, unique RFLP patterns (130+ 310 bp bands) were obtained for 6 strains of Nocardia (En28, En39, En43, En46, En47, and En49). In these strains, the MspI restriction enzyme produced restriction patterns consisting of three bands (three strains) or four bands (four strains) and HinfI enzymes produced restriction patterns consisting of two bands in these 6 isolates (Table 1; Figure 2). Eventually, after analyzing the RFLP band patterns generated by each of the three enzymes, we detected new RFLP patterns “A” and “B”. We performed the 16S rRNA gene sequences (1500bp) for all the isolates of Nocardia, especially for strains that gave the new RFLP patterns (See Table 1). The gene Bank accession numbers of the 16S rRNA gene for Nocardia isolates in our study are given in Table 2. Based on phenotypic test results in our previous report (17), isolate En28 was identified as N. africana. After the analysis of PCR - RFLP of the hsp65 gene, we observed a new pattern “A” (BstEII, 130/310; MspI, 120/140/180; HinfI, 190/250). This isolate after conducting the sequence analysis of 16S rRNA gene was 100% similar to N.ignorata DSM 44496. The RFLP profile of the hsp65 gene for N. ignorata according to the report of Rodriguez - Nava et al. was BstEII, 80/320; MspI, 130/120/115/70; HinfI, 190/250 (14). After performing the BLAST comparison, isolates En46 and En47 (whit RFLP pattern B) showed the greatest similarity to N. cummidelens HBUM174688 (En 46; similarity 99.66% with a difference in 5 nucleotides, En47; similarity 99.93% with a difference in 1 nucleotide), when compared to the NCBI nucleotide sequence (25). Isolates En39 and En43 had a similar RFLP pattern “B” (BstEII, 130/310; MspI, 70/110/120/140; HinfI, 190/250), which, after sequencing of 16S rRNA gene, were identified as N. otitidiscaviarum and N. soli, respectively (See Table 1). Steingrube et al. (1995) reported various patterns for N. asteroides and N. otitidiscaviarum by MspI for initial digestion and then restriction with BsaHI for an amplified 439 - bp segment of the 65 - kDa heat shock protein gene (DNA amplification and restriction) (30). According to previous studies, the RFLP pattern of the hsp65 gene was reported for N. ignorata as BstEII, 80/320; MspI, 130/120/115/70; HinfI, 190/250, N. soli and N. cummidelens as BstEII, 80/320; MspI, 130/120/115/70; HinfI, 190/250, N. asteroids type IV, N. cyriacigeorgica, and N. neocaledoniensis as BstEII, 440; MspI, 180/145-130/120 - 115; HinfI, 440 (14). In this study, we obtained new RFLP patterns for N. cummidelens, N. ignorata, and N. soli (See Table 2). It is possible that some Nocardia spp. contained two different hsp65 restriction - endonuclease fragment patterns (or more than two). Therefore, the hsp65 restriction - endonuclease fragment pattern results for N. ignorata, N. soli, and N. otitidiscaviarum differed from those of other research (14). The 16S rRNA gene sequencing showed that strains En8 and En39 both belonged to N. otitidiscaviarum, while strain En8 had the expected hsp65 restriction - endonuclease fragment pattern D and strain En39 had a new pattern B. The RFLP pattern obtained in the isolate En49 (RFLP pattern B:BstEII, 130/310; MspI, 70/110/120/140; HinfI, 190/250) due to the sequence similarity of the 16S rRNA gene (Figure 3, similarity 99.06% to N. coubleae OFN N11) was considered as N. coubleae (See Table 2). It is noteworthy that, any pattern for the RFLP of hsp65 gene was not identified for N. coubleae until now. Hence, determining the hsp65 gene patterns in various species of Nocardia can be helpful to get a more accurate diagnosis. In addition to the identification of Nocardia spp., the PCR - RFLP and sequencing techniques could be used for detection and differentiation of unusual species of Nocardia and distinguish Nocardia and Mycobacterium species from each other (30). Based on the findings of previous published papers and the results of the present study, the same RFLP pattern of hsp65 gene could be present in several different Nocardia species. Similarly, the phenotypic properties may be different even in Nocardia isolates that belong to the same species. PCR - RFLP analysis targeting hsp65 is a rapid method for identification of Nocardia species if the pattern of all species was determined in databanks of NCBI. Therefore, it is recommended that RFLP - hsp65 profiles be determined to create a comprehensive database for all discovered species of Nocardia.
4.1. Conclusions
We obtained two new PCR - RFLP profiles of hsp65 gene for Nocardia isolates from soil. Strains that exhibited new RFLP profiles should be further analyzed by sequence - based methods such as full sequences of 16S rRNA gene.