1. Background
Helicobacter pylori is a gram-negative, spiral-shaped bacterium first discovered by Robin Warren and Barry Marshall in 1982 (1). This bacterium is considered one of the most successful pathogens in human infection, affecting 50 - 60% of the global population, approximately 4.4 billion people as of 2015 (2, 3). Although H. pylori primarily resides in the stomach, it can also extend to the distal esophagus or proximal duodenum in cases of gastric metaplasia (4). Its unique characteristics allow it to alter the gastric environment and reduce acidity (5). Helicobacter pylori infection is strongly associated with many digestive system diseases, such as gastritis, dyspepsia, peptic ulcers, duodenal ulcers, lymphoma of mucosa-associated lymphoid tissue (MALT), and gastric adenocarcinoma (6).
The prevalence of infections caused by this pathogen varies by region. For instance, in Latin American countries, the prevalence is high at 75.83%, compared to 39.6% in Japan and 17.1% in the United States (7). In Iran, serological methods indicate a 69% prevalence rate across the general population, rising to 89% among individuals over 40 years old in high-prevalence regions, such as the northwest, as determined by serological and pathological methods (8). Helicobacter pylori infection, along with factors, such as smoking and NSAID use, is reported to contribute to 3.3% to 4.4% of gastric and duodenal ulcers (9).
Several diagnostic methods exist for detecting H. pylori infections, including endoscopic biopsy, urea breath test, serological tests, stool antigen tests, polymerase chain reaction (PCR), and enzyme-linked immunosorbent assay (ELISA) (10). Genetic factors play a significant role in diagnosing and treating stomach diseases, as identifying the Helicobacter gene can enhance the accuracy of clinical diagnosis (11). Among molecular techniques, the PCR method is particularly prevalent, as it is used to amplify H. pylori DNA from fecal samples (12). Helicobacter pylori possesses several strains, which allow it to persist chronically within host epithelial cells (13). For optimal testing, stool samples should be analyzed promptly; however, if stored at temperatures ranging from -5 to 25°C, they can remain viable for testing up to 7 days (14).
The ELISA test utilizes components of the body’s immune system, such as antibodies and specific chemical agents, to detect immune responses to foreign entities like infectious microbes (15). However, while serological diagnostic methods are useful, they tend to have low specificity. Moreover, serology results do not necessarily confirm an active infection. Consequently, this technique is not reliable for confirming the eradication of an infection (16).
Conversely, PCR is a highly accurate method that employs multiple gene targets to detect H. pylori (17). While both ELISA and PCR methods exhibit high sensitivity compared to other diagnostic techniques, their comparative accuracy requires further investigation (18).
2. Objectives
The primary objective of this study was to assess and compare the diagnostic accuracy of ELISA and PCR methods in detecting H. pylori infections in patients presenting with gastric issues. The significance of this study lies in its potential to provide a quicker and more accurate diagnosis of the infection, thereby improving the management and treatment of gastrointestinal diseases. This is crucial for facilitating timely and effective therapeutic interventions.
3. Methods
This diagnostic study was conducted on 70 patients referred to Khurshid Private Laboratory in Tehran from July to September 2022. Inclusion criteria included individuals aged 18 years or older exhibiting gastric disease symptoms, such as ulcers or gastric adenocarcinoma. Participants should not have taken antibiotics or proton pump inhibitors recently, should have no history of gastric surgery, and must be willing to undergo diagnostic procedures. They should also maintain stable health without any severe illness. Exclusion criteria included recent use of antibiotics, NSAIDs, bismuth, corticosteroids, or PPIs within the past two weeks, a history of upper GI bleeding, renal failure, pregnancy, chronic liver disease, gastric carcinoma, being under 18 years old, or having diabetes. Prior to sample collection, patients completed a questionnaire regarding their personal and medical history, ensuring confidentiality was maintained during data handling.
3.1. Sampling
After the patients completed the questionnaire and consent forms, blood specimens were collected using a sterile 10 cc syringe, and stool specimens were collected under sterile conditions. The serum samples were then sent to the immunology laboratory for the extraction of IgG and IgA using the ELISA test. The stool samples were sent to the molecular laboratory for the extraction of H. pylori DNA using the PCR technique.
3.2. Immunology Test
The levels of IgA and IgG antibodies were measured using the indirect ELISA method with an ELISA kit provided by the Pishtaz Teb Company. IgG and IgA titers were considered positive for values of 10 or above, and negative for values below 10.
3.3. Molecular Test
The homogenized fecal sample DNA extraction temperature was set at -70°C. DNA analysis of the fecal sample was performed using the Salting Out method with the DNP kit from Cinnagen. DNA was extracted quantitatively (1.6 < OD < 1.9) and qualitatively using PCO3 (5'-ACACAACTGTGTTCACTAGC-3') and PCO4 (5'-CAACTTCATCCACGTTCACC-3') primers, capable of amplifying a section of the human β-globulin gene.
3.4. Initiation of Synthesized Primers and Probes
The primers and oligonucleotide probes were synthesized according to the DNA/RNA synthesis model 394. Primer 93275 (5’- AAGCTTTTAGGGGTGTTAGGGGTTT-3’) and primer 93276 (5'-AAGCTTACTTTCTAACACTAACGC-3’) were used, which have been previously employed to detect H. pylori infection (19), by targeting the sequence of the ureC gene (GeneBank numbers X57132, EMBL, M60398). These primers amplify a DNA fragment of 294 base pairs. The probe used (5'-CGATTGGGGATAAGTTTGTGA-3') was designed to identify the ureC gene by binding to nucleotides 137 - 158 of the amplified fragment. The probe was biotinylated according to a new method (20).
3.5. Polymerase Chain Reaction Assay Design
The reactions were conducted in a total volume of 50 microliters using an Eppendorf thermocycler. The reaction mixture contained 0.4 micromoles of each primer, 0.2 micromoles of each deoxynucleotide triphosphate, and 0.01 micromole of Digoxigenin (DIG-dUTP) (Boehringer Mannheim GmbH, Mannheim, Germany), along with the reaction buffer (100 mM Tris-HCl, 15 mM MgCl2, 500 mM KCl). Additionally, 0.01 units of Taq polymerase (Boehringer) were added before amplification, followed by the addition of mineral oil. Finally, 10 microliters of sample DNA were added to the reaction mixture. The thermal cycling conditions were set according to the temperature profile in Table 1. After amplification, PCR products were visualized by electrophoresis for 2 - 4 minutes using an agarose gel stained with 0.5 - 1 μg/mL Ethidium bromide, and the results were photographed under ultraviolet light.
| Function | Temperature, °C | Time | Number of Cycles |
|---|---|---|---|
| PCR | 35 | ||
| Denaturation | 94 | 30 s | |
| Annealing | 54 | 30 s | |
| Extension | 72 | 1 m | |
| Final amplification | 72 | 10 m | 1 |
3.6. Data Analysis
Qualitative variables were described using frequency and percentage, while quantitative variables were described using the mean and standard deviation. The Chi-square test was used to compare qualitative variables, and the t-test (for variables with a normal distribution) or the Mann-Whitney test (for non-normally distributed variables) was used to compare quantitative variables. To compare the average levels of IgA and IgG across different age groups, ANOVA and the Kruskal-Wallis test were applied. Additionally, to determine the optimal cut-off points for IgA and IgG in diagnosing H. pylori infection, sensitivity and specificity were calculated, and the receiver operating characteristic (ROC) curve was used. Data analysis was conducted using Stata version 14 software, and a P-value of < 0.05 was considered statistically significant.
4. Results
A total of 70 patients with stomach diseases participated in the study. The average age of the patients was 43.02 ± 15.49 years (age range: 13 to 83 years). Of the patients, 51.43% (36 individuals) were male, and 48.57% (34 individuals) were female. Based on PCR test results, the prevalence of H. pylori was 24.29%. The mean levels of H. pylori IgA and H. pylori IgG in patients were 5.83 ± 5.12 and 47.01 ± 56.66, respectively. According to PCR results, the prevalence of H. pylori was 22.22% in males and 26.47% in females. The prevalence was 25% in individuals under 25 years old, 18.42% in those aged 25 - 50 years, and 33.33% in individuals older than 50 years. Among patients, 52.94% of those with nausea, 68.18% of those with flatulence, and all individuals with a history of malignancy tested positive for H. pylori. Therefore, a significant relationship was observed (Table 2). The average age of individuals with H. pylori infection was higher than that of those without the infection, but this difference was not statistically significant (P = 0.24).
| Variables | Positive (n = 17) | Negative (n = 53) | P-Value b |
|---|---|---|---|
| Gender | 0.67 | ||
| Male | 8 (22.22) | 28 (77.7) | |
| Female | 9 (26.4) | 25 (73.3) | |
| Age (y) | 0.41 | ||
| < 25 | 2 (25) | 6 (75) | |
| 25 - 50 | 7 (18.42) | 31 (81.5) | |
| > 50 | 8 (33.3) | 16 (66.6) | |
| Nausea | 0.002 | ||
| Yes | 9 (52.4) | 8 (47) | |
| No | 8 (15) | 45 (84.5) | |
| Flatulence | < 0.001 | ||
| Yes | 15 (68.1) | 7 (31.8) | |
| No | 2 (4.1) | 46 (95) | |
| Loss of appetite | 0.57 | ||
| Yes | 5 (29.4) | 12 (70.6) | |
| No | 12 (22.6) | 41 (77.6) | |
| History of gastrointestinal malignancy | < 0.001 | ||
| Yes | 5 (100) | 0 | |
| No | 11 (17.2) | 53 (82.8) |
a Values are expressed as No. (%).
b Using the chi-square test.
4.1. Receiver Operating Characteristic Curve Results
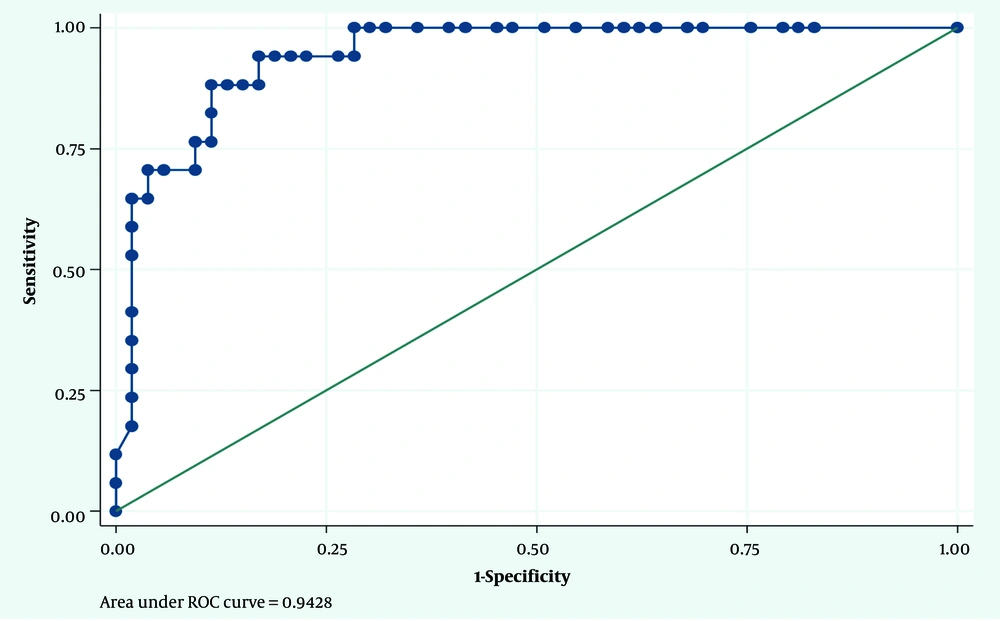
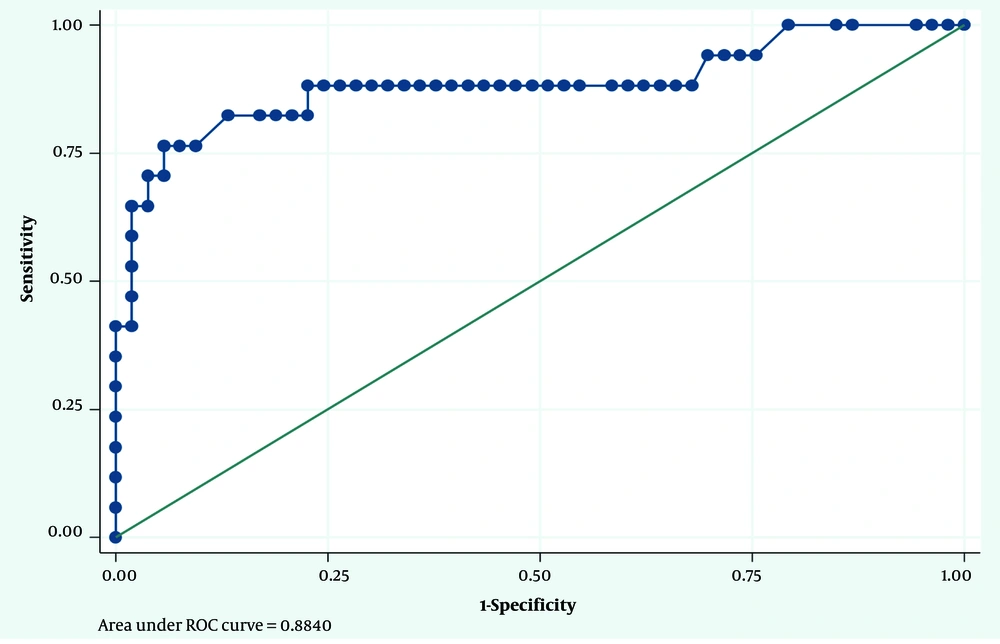
Considering the PCR results as the gold standard, the optimal cut-off point for IgA in diagnosing H. pylori infection was 7.15 mg/dL. The sensitivity and specificity for this cut-off point were 88% and 89%, respectively. The area under the ROC curve (AUC) was calculated as 0.94 (Figure 1). Similarly, considering the PCR results as the gold standard, the optimal cut-off point for IgG in diagnosing H. pylori infection was 95.70 mg/dL. The sensitivity and specificity for this cut-off point were 76% and 94%, respectively. The AUC was calculated as 0.88 (Figure 2).
5. Discussion
The polymicrobial nature of gastrointestinal diseases is clinically significant (21). Specifically, H. pylori infection is a primary cause of gastritis and gastric ulcers in humans and is also considered a risk factor for gastric cancer. According to the International Agency for Research on Cancer, H. pylori infection is classified as a group I carcinogen in humans (22). Over 50% of the world’s population is infected with H. pylori, although the infection rate varies between countries (23), ranging from 85% to 95% in developing countries and 30% to 50% in developed countries (24). Early diagnosis is crucial for effective management and the appropriate prescription of antibiotics to eradicate the pathogen and prevent its complications. There are various invasive and non-invasive techniques for diagnosing H. pylori infection (25), including microbiological culture (26), rapid urease test (RUT), biopsy-based PCR (27), urea breath test (UBT) (28), stool antigen tests (SAT) (29), and serological assessments (30). Stool specimens are used in molecular tests for the non-invasive detection of H. pylori DNA (31). The ureC PCR method targets the urease C (ureC) gene. It was once incorrectly believed that the ureC gene was associated with urease formation (32). However, it was later discovered to be involved in the production of phosphoglucosamine mutase, which plays a role in bacterial cell wall synthesis, and it is now known as the glmM gene (33).
In our study, the prevalence of H. pylori in patients, as determined by PCR, was found to be 24.29%, with a prevalence of 22.22% in men and 24.67% in women. A previous study conducted in 2019 on 350 stool samples, comprising 37% women and 63% men aged between 1 and 68, reported that 41% of the samples were positive for H. pylori using the PCR method (34). Another study, carried out in 2022 on 124 samples, found that 81.52% were positive for H. pylori via PCR (28). Additionally, a 2022 study aimed at detecting H. pylori in patient stool samples using the multiplex urea PCR method reported a 65% positivity rate (35).
Helicobacter pylori antibodies, including IgA and IgG, can be detected in stool, blood serum, and saliva through serological methods such as ELISA (27, 29). In our study, the mean levels of H. pylori IgA and IgG antibodies in patients were 5.83 ± 5.12 and 47.01 ± 56.66, respectively. A 2020 study focusing on identifying H. pylori in cases of gastric ulcers, conducted on 137 samples, found that 29.2% were positive for IgA and 71.5% for IgG based on ELISA results (28). Furthermore, a 2022 study aimed at diagnosing H. pylori in patients with peptic ulcers using blood samples revealed that 52% of the samples tested positive via ELISA (36).
These findings highlight the variability in H. pylori prevalence across different populations and studies, emphasizing the necessity of employing accurate diagnostic methods to ensure effective detection and subsequent treatment of H. pylori infections. The use of both PCR and ELISA can provide complementary insights, aiding in better management of associated gastrointestinal diseases.
The choice of diagnostic technique for H. pylori infections is influenced by factors such as sensitivity, specificity, cost considerations, and clinical status (10). Each testing method presents unique limitations, advantages, and disadvantages depending on clinical circumstances and patient history (37). Polymerase chain reaction is noted for its high sensitivity and specificity, exceeding 95%, compared to other conventional methods (38). In our study, using PCR results as the gold standard for identifying H. pylori, the sensitivity and specificity for detecting H. pylori IgA were 88% and 89%, respectively, while for IgG, they were 76% and 94%.
A 2021 study that diagnosed H. pylori infection using both invasive and non-invasive methods in patients with digestive disorders reported that 81% of samples were positive via the qPCR method, whereas 53% were positive via the ELISA method. In this study, the sensitivity and specificity of the IgG ELISA test were found to be 73.5% and 85.3%, respectively, with PCR identified as the method with the highest accuracy and sensitivity (25).
Further, a 2019 study analyzing diagnostic methods for identifying H. pylori reported that, based on the PCR method, 51.9% of the 102 samples tested positive, compared to 30.4% positivity for IgA using ELISA. The sensitivity and specificity reported were 66.8% and 75%, respectively (39). Another comparative study in 2018 between real-time PCR and ELISA found that, for 87 samples, the PCR method had a detection rate of 81.6%, while the ELISA method had a sensitivity of 87% and specificity of 60%, respectively (40).
These studies underscore the critical link between H. pylori infections and various digestive system diseases, such as gastric ulcers, duodenal ulcers, gastric flatulence, and gastric adenocarcinoma (41). In our study, a significant correlation was observed between clinical symptoms, such as nausea and flatulence, and a history of gastrointestinal malignancy, with positive Helicobacter PCR results. This finding emphasizes the importance of selecting an appropriate diagnostic approach for H. pylori to ensure accurate detection and effective management of gastrointestinal diseases linked to this bacterium.
In a study conducted in 2018, the prevalence of H. pylori infection was reported as 64.39%, and all patients with abdominal pain, frequent belching, and abdominal bloating tested positive for H. pylori infection (42). In a 2019 study of 158 samples, H. pylori was found to be associated with stomach cancer, a history of malignancy, family history, and age (43). Additionally, in the Balabel study conducted in 2022, bloating, nausea, and heartburn were most commonly associated with H. pylori infection (44).
As seen in the results of our study, it is important to consider symptoms such as nausea, bloating, and a history of malignancy in individuals with H. pylori. Due to the high sensitivity and specificity of the PCR method, this test has been introduced as the gold standard for detecting H. pylori in stool samples. However, the sensitivity and specificity of the ELISA method were also high, suggesting that these two methods should be used together in the diagnosis of Helicobacter infection.
5.1. Limitations
This study compares the diagnostic accuracy of ELISA and PCR for detecting H. pylori, but it faces notable limitations. The small sample size of 70 participants may not represent a broader population, potentially affecting the generalizability of the results. Relying on both stool and serum samples poses challenges in maintaining consistent quality, risking DNA degradation and compromised results. Conducted in a single Tehran laboratory, the study may be subject to location-specific biases. The short three-month timeframe may not capture seasonal variations affecting H. pylori prevalence, highlighting the need for expanded research with larger sample sizes and broader geographic and temporal coverage.
5.2. Conclusions
This study highlights the critical importance of accurately diagnosing H. pylori infections, which are strongly linked to various gastrointestinal diseases, such as gastritis, gastric ulcers, and gastric cancer. By comparing the diagnostic accuracy of ELISA and PCR methods, the research provides valuable insights into optimizing detection strategies for H. pylori. Polymerase chain reaction demonstrates higher sensitivity and specificity compared to ELISA, making it a more reliable gold standard for diagnosing this infection. Given that the prevalence of H. pylori varies significantly across populations and the infection is classified as a group I carcinogen, timely and accurate diagnosis is essential for effective management and treatment. Establishing optimal cut-off points for IgA and IgG, as identified in this study, allows for enhanced diagnostic precision, which can lead to better-targeted therapeutic interventions and improved patient outcomes. This research underscores the necessity of selecting appropriate diagnostic techniques, considering the clinical context, to ensure the effective management of H. pylori-related gastrointestinal diseases.