1. Background
Today, one of the most pressing issues threatening global health systems is infection, particularly treatment-resistant forms, which lead to substantial and escalating costs for healthcare systems (1-3). Nosocomial infections (NIs), especially those caused by antibiotic-resistant strains, are a serious concern in healthcare settings and impose considerable financial burdens on national healthcare sectors.
Escherichia coli bacteremia (ECB) is a major infection in both humans and animals (4, 5). Escherichia coli is a genus of gram-negative bacteria and a member of the Enterobacteriaceae family (4, 5). In healthcare settings, E. coli is a primary and common cause of NIs that can cause disease in humans (4, 6). The primary reservoirs of E. coli are healthy individuals and animals (7). Among the most notable side effects of E. coli infections are diarrhea, stomach cramping, pain, tenderness, nausea, and vomiting (7-10).
Medical reports have indicated that E. coli is a leading cause of urinary tract infections in children and women (11). As one of the most common pathogens responsible for infection, the use of antibiotics has grown significantly, which can lead to antibiotic resistance (11). Reports from health organizations in less-developed and developing countries indicate that Enteropathogenic E. coli (EPEC) is a leading cause of diarrhea in children (12). In recent decades, one of the most critical challenges facing global healthcare systems has been the excessive and inappropriate use of antibiotics, resulting in resistance to commonly used treatments (12).
The primary causes of NIs include exposure to infected medical or diagnostic tools, contaminated environments, contact with medical staff, and interactions between patients and healthcare personnel (13-15). Various antimicrobial agents, including clindamycin, glycopeptides, beta-lactam antibiotics resistant to penicillinase, sulfonamides, and aminoglycosides, are used to treat E. coli-caused diseases (16-19). Methicillin-resistant E. coli is one of the most common nosocomial pathogens and a leading cause of NIs, with its prevalence steadily increasing worldwide in recent decades (20). One of the main contributors to the rise in antibiotic resistance is the widespread and excessive use of antibiotics in food production and industries such as livestock, poultry, dairy, and agriculture (21). Symptoms of antibiotic resistance and the overuse of antibiotics include genetic transmission within populations and the spread of antibiotic-resistant genes in cultured specimens (22). Pathotypes of E. coli, including Enterohemorrhagic E. coli (EHEC), are significant causes of both enteric and extra-intestinal diseases. Enterohemorrhagic E. coli strains are particularly known as major agents of diarrhea in communities with low levels of welfare and financial resources (23). Although some governments have implemented restrictions on the use of antibiotics in animals, misuse of antibiotics continues to be widespread throughout many countries (4).
Increased hospital costs, extended healthcare expenditures, prolonged hospitalization, and high mortality rates are significant disadvantages of NIs (24). Healthcare personnel involved in the control and prevention of infectious diseases in hospitals and health centers must measure the prevalence of these microorganisms and implement protection programs to prevent bacterial spread. To achieve this goal, understanding the antibiotic resistance model and the factors and genes influencing resistance is crucial for infection control programs (4).
Colistin is recognized as a last-resort treatment for multidrug-resistant gram-negative pathogens. However, the mobilized colistin resistance (mcr-1 gene) gene is increasingly being identified in bacterial isolates from humans, animals, farms, food, and the environment (25). In 2015, the plasmid-mediated mcr-1 gene was first described in E. coli and Klebsiella pneumoniae (26). Recently, Partridge et al. identified fourteen allelic variants of mcr-1 (27).
Antibiotic-resistant infections are increasing globally. For example, the WHO reported that approximately 700,000 deaths occur annually due to drug-resistant infections. This number is projected to rise, potentially reaching 10 million deaths per year by 2050 if current trends continue. Multi-drug-resistant organisms, particularly gram-negative bacteria such as E. coli and Klebsiella pneumoniae, are major contributors to this crisis. Colistin, a polypeptide antibiotic, is often considered a last-resort option for treating infections caused by multidrug-resistant gram-negative bacteria. Its use has surged due to ineffective alternatives, particularly in critical care settings. The mcr-1 gene encodes an enzyme that modifies the bacterial cell membrane, reducing colistin's efficacy. This gene can confer resistance not only to colistin but also to other antibiotics, complicating treatment protocols. The significance of the mcr-1 gene highlights a critical aspect of the antibiotic resistance crisis, particularly in the context of colistin as a last-resort antibiotic. The global spread of this gene and the associated resistance it confers underline the urgent need for coordinated action to combat antibiotic-resistant infections.
Given that microbial diagnostic and sensitivity tests lack the ability to diagnose bacteria and assess resistance to E. coli antibiotics, there is a growing need for more accurate methods, such as polymerase chain reaction (PCR), which offer greater specificity (21). The conventional PCR method is more sensitive than the culture method (21).
2. Objectives
This study aimed to determine the pattern of E. coli antibiotic resistance (ECAR) and identify the mcr-1 gene in isolates from non-clinical samples using conventional PCR in Asadabad, 2017.
3. Methods
3.1. Study and Samples
In this descriptive cross-sectional study, the sample size was calculated to be 265 samples based on a recent study. This included 79 clinical samples (human urine) and 186 animal feces samples, which were collected aseptically in Asadabad city, Hamadan province, Iran, from 15 August to 25 December 2018 (21, 28). The sampling method involved clinical isolates of urine samples. The animal feces were collected from poultry (chickens, turkeys) and livestock farms (cattle, sheep, goats).
First, all samples were cultured on MacConkey agar (MA) and Eosin methylene blue (EMB) agar for 48 hours at 37°C. Then, all colonies typical of E. coli were transferred to nutrient agar for 24 hours at 37°C. A 95% confidence interval (95% CI), a standard error of 5% or less, and frequency data were used in this study to estimate the required sample size for assessing ECAR and the presence of the mcr-1 gene in isolates from non-clinical samples (28).
3.2. Examining Antibiotic Sensitivity
In this study, 30 clinical E. coli isolates were collected from laboratory urine samples, and 70 isolates were obtained from animal feces from poultry and livestock farms in Asadabad in 2017. The antibiotic susceptibility testing of the isolated E. coli was performed using the standard disk diffusion method (Bauer-Kirby) on Mueller Hinton agar (MHA) medium, as recommended by the Clinical and Laboratory Standards Institute (CLSI-2017) guidelines. Six antibiotic discs were used, including nalidixic acid (30 μg), trimethoprim-sulfamethoxazole (25 μg), ampicillin (10 μg), ciprofloxacin (5 μg), gentamicin (5 μg), and colistin (10 μg), and the plates were incubated at 35°C for 16 - 18 hours in 5% CO2. Based on the diameter of the inhibition zones, the isolated E. coli were classified into two categories: Sensitive and resistant (resistant and intermediate E. coli), according to the CLSI guidelines. After DNA extraction, a PCR test was performed to detect the mcr-1 gene in the E. coli isolates.
3.3. Molecular Test Procedure
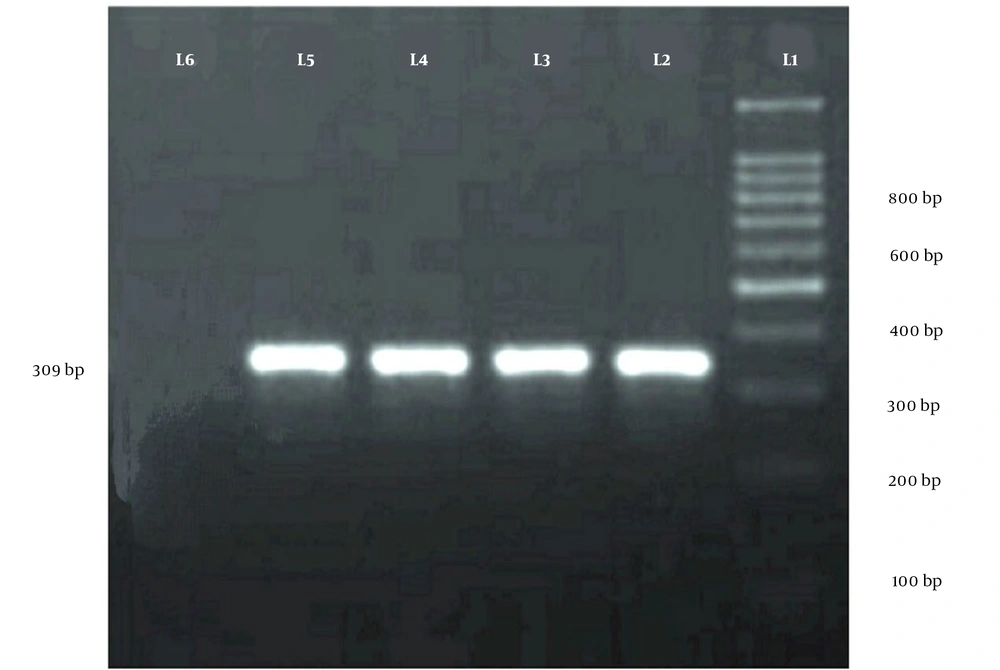
In the present study, the mcr-1 gene in E. coli cultured samples was diagnosed and confirmed using PCR. In the first stage, commercial DNA extraction kits (Sina Clon, Iran) were used to extract chromosomal DNA from all isolates.
Briefly, a pure colony of fresh bacterial culture was inoculated into microtubes containing a pre-lysis medium. After incubating at 55°C for 30 minutes, lysis buffer and precipitation solution were added. The samples were then washed three times at 13,000 rpm for 1 minute using a pipette and centrifuge. After adding 30 μL of elution buffer, the sample was incubated at 65°C for 3 - 5 minutes, followed by centrifugation. The supernatant, containing the DNA, was transferred to a sterile microtube and stored at -20°C until the PCR reaction.
The quality and quantity of the extracted DNA were assessed using a spectrophotometer and agarose gel electrophoresis. Optical absorbance at wavelengths of 280 nm and 260 nm was used to determine the concentration and purity of the samples. A DNA purity ratio between 1.6 and 1.8 was considered appropriate.
For final confirmation, PCR was performed using primers specific for the mcr-1 gene (Table 1). The PCR reaction was conducted in a 25 μL volume containing 12.5 μL of master mix, 1 μL of forward and reverse primers, 1 μL of DNA sample, and 9.5 μL of distilled deionized water. The thermal cycling conditions were as follows: An initial denaturation at 95°C for 10 minutes, followed by 34 cycles of 95°C for 45 seconds, 50°C for 45 seconds, and 72°C for 40 seconds. The final extension was carried out at 72°C for 7 minutes. The PCR reaction was performed in a thermocycler (BioRad, USA). Escherichia coli ATCC 25423 and ATCC 25923 were used as control strains.
| Gene Targets | Primer Sequences (5' to 3') | Amplicon/Product Size (bp) |
|---|---|---|
| mcr-1 | CLR5-F CGGTCAGTCCGTTTGTTC | 309 |
| CLR5-R CTTGGTCGGTCTGTAGGC |
The procedure for examining antibiotic sensitivity, known as the antibiogram test, was performed using the standard disk diffusion method (Bauer-Kirby) on Mueller Hinton agar medium. This test was conducted according to the disk diffusion method and the guidelines of the National Committee for Clinical Laboratory Standards (CLSI) (9). Five different antibiotic discs were used: Gentamicin (10 mg), trimethoprim-sulfamethoxazole (10 mg), ciprofloxacin (5 μg), oxacillin (1 μg), and erythromycin (15 mg) (Table 2).
| Antibiotics | Clinical Samples | Livestock Examples | ||
|---|---|---|---|---|
| Resistance | Sensitive | Resistance | Sensitive | |
| Ampicillin | 13 (43.43) | 17 (56.66) | 56 (80) | 14 (20) |
| Trimethoprim sulfamethoxazole | 10 (33.33) | 16 (53.34) | 49 (70) | 21 (30) |
| Nalidixic acid | 10 (33.33) | 20 (66.67) | 56 (80) | 14 (20) |
| Ciprofloxacin | 14 (46.66) | 20 (66.67) | 63(90) | 7 (10) |
| Gentamicin | 7 (23.33) | 23 (76.67) | 7 (10) | 63 (90) |
| Colistin | 5 (16.66) | 25 (83.34) | 10 (14.28) | 60 (85.72) |
a Values are expressed as No. (%).
Afterward, the isolated strains were identified and confirmed using standard biochemical tests, including Gram stain, oxidase test, catalase, DNase, coagulase, and mannitol fermentation. The confirmed isolates were stored in a freezer at -20°C for subsequent experiments. All 31 strains of Staphylococcus aureus were confirmed through phenotypic tests. After obtaining informed consent, all relevant information was entered into the statistical software.
3.4. Statistical Analysis
The analyses were conducted using IBM SPSS Statistics for Windows, version 24. Descriptive statistical measures, including mean, standard deviation, and percentages, were used to describe the data. The estimated prevalence rates were presented with a 95% CI. To determine whether there was a significant difference in the frequency of isolates among the three types of samples, the chi-square test was used. To analyze the antibiotic resistance model of E. coli isolates, clinical and animal samples were utilized, and the number and percentage of resistant and sensitive isolates were presented for each antibiotic. To evaluate the significance, the P-value was calculated from the chi-square test. A P-value of < 0.05 typically indicates a statistically significant difference between groups.
4. Results
Out of 265 samples (clinical and fecal), 37.97% (30/79) of clinical samples and 37.63% (70/186) of animal feces showed growth of E. coli. The highest number of E. coli isolates (71.48%; 50/70) came from livestock farm feces. The antimicrobial susceptibility test of E. coli isolates from clinical samples showed that the highest resistance (46.66%) was against ciprofloxacin, and the lowest resistance (16.66%) was against colistin. Among the study samples, 36.66% of the 30 clinical samples were mcr-1 gene carriers, and 89.12% of ciprofloxacin-resistant isolates were diagnosed as mcr-1 positive. Of the isolates containing the mcr-1 gene, 72.72% were clinical isolates, and 82.35% were ciprofloxacin-resistant isolates from animals (Figure 1). The test for isolates collected from animal samples showed that the highest resistance (90%) was against ciprofloxacin, and the lowest (10%) was against gentamicin. Additionally, the results demonstrated that 36.66% (11/30) of E. coli isolates from clinical urine samples and 24.28% (17/70) of E. coli isolates from animal feces possessed the mcr-1 gene.
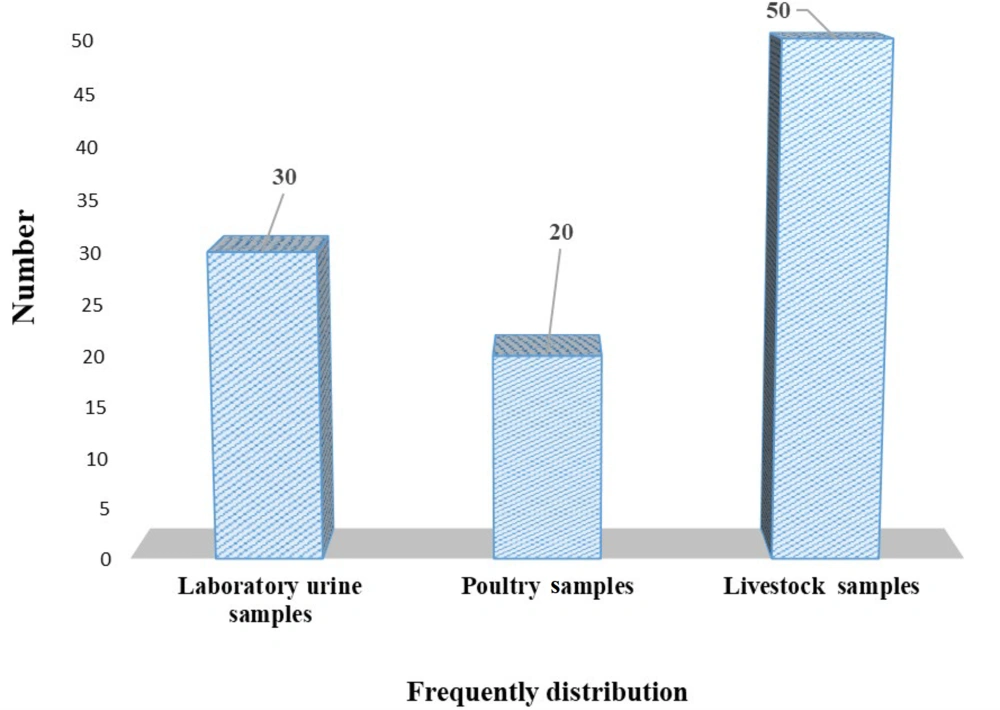
The results of this study showed that out of 79 clinical samples, 30 isolates of E. coli were identified from laboratory urine samples. Additionally, from 186 animal samples, 20 poultry samples and 50 livestock samples were identified (Figure 2).
The pattern of antibiotic resistance of E. coli isolated from patients with urinary tract infections in the studied hospitals is shown in Table 2. Out of 79 clinical samples, 30 isolates of E. coli were identified from laboratory urine samples. Additionally, from 186 animal samples, 20 poultry samples and 50 livestock samples were identified in this study (Table 2).
The highest percentages of resistance in clinical and animal samples were observed for the antibiotic ciprofloxacin, with 46.66% and 90% resistance, respectively. The lowest percentages of resistance were observed for gentamicin, with 23.23% in clinical samples and 10% in animal samples.
5. Discussion
Escherichia coli is the main species of facultative anaerobe in the gastrointestinal tract of humans and animals. While it is typically a harmless microbe, it is still known as one of the main risk factors in causing NIs (29, 30). Many studies have shown that improper selection and misuse of antibiotics can lead to antibiotic resistance, making the treatment of bacterial infections difficult. This issue is one of the most significant threats to global health (30).
Detection of the mcr-1 gene in clinical and non-clinical E. coli isolates in healthcare settings, such as hospital wards, is essential because these environments can act as potential sources of microorganisms. According to the results obtained from PCR analysis, 36.66% of E. coli isolates from 30 clinical samples were mcr-1 gene carriers. Among ciprofloxacin-resistant isolates, 89.12% were mcr-1 positive. Among the isolates containing the mcr-1 gene, 72.72% were clinical isolates, while 82.35% were ciprofloxacin-resistant animal isolates.
Doosti et al. in Farsan, Iran, evaluated E. coli strains isolated from urinary tract infections in children and women and their antibiotic resistance patterns (11). They reported that, in the antimicrobial resistance test, the highest sensitivity was observed to ciprofloxacin (31.46%) and the highest resistance to ampicillin (78.95%). According to the PCR results, they determined that the most common resistance genes were CTX, with the lowest incidence of SHV-B resistance (11).
In another study, Karami et al. in Hamadan, Iran, evaluated EPEC strains in children with diarrhea and their antibiotic resistance patterns (12). Based on their results, the rate of multi-drug resistance was 30%, with the highest resistance seen in cefpodoxime (97%). This study also showed that the most effective antibiotics were imipenem and ciprofloxacin (12).
In 2017, Hosseinpour et al. in Hamadan studied the types of E. coli isolated from poultry and food based on phenotypic and molecular tests (21). Their results indicated that the maximum antibiotic resistance was observed for nalidixic acid and ampicillin, with resistance rates of 88.4% and 76.8%, respectively (21).
Bista et al., in a 2019 study in Nepal, investigated colistin-resistant E. coli and the presence of the mcr-1 gene in chicken liver samples (4). They reported that 53.3% of the samples showed growth of E. coli. The study found that birds less than 40 days old had the highest frequency of E. coli isolation. In antimicrobial susceptibility testing (AST), 95.1% and 82.6% of the E. coli isolates were resistant to tetracycline and ciprofloxacin, respectively (4).
Previous studies have reported varying prevalence rates of the mcr-1 gene in E. coli isolates from clinical and agricultural settings. For instance, research in Europe often shows lower prevalence rates (typically below 5%), while studies in Asia, particularly in China, report significantly higher rates (above 20%). Comparing these rates with the findings of this study can provide insight into patterns related to local agricultural practices, antibiotic use, and public health interventions. The variation in prevalence may also result from differences in sample sources. Studies focusing on human clinical isolates may show different resistance patterns compared to those from livestock or environmental sources. The geographical context and temporal factors are also critical in resistance profiles. Regions with intensive agriculture and high antibiotic use in livestock are more likely to report elevated mcr-1 prevalence. Conversely, studies reflecting the impact of stricter antibiotic regulations may show declining prevalence over time, especially in areas where less stringent controls were previously in place.
The use of colistin as an antibiotic in agricultural settings is a particular concern. In regions where colistin is commonly used for growth promotion or disease prevention in livestock, higher prevalence rates of mcr-1 are often reported. If the current study finds high resistance in livestock, it may correlate with historical or current agricultural practices. Additionally, variations in infection control practices between hospitals and farms can influence the spread of resistant strains. Increased biosecurity measures in some regions may lead to lower transmission, while lax practices in others may facilitate the spread of resistant bacteria.
In a similar study, Zhong et al. in Changsha examined patients with bloodstream infections and their association with the mcr-1 gene (7). They reported that 2.1% of the 144 E. coli isolates were positive for the mcr-1 gene. They also found that the three mcr-1-positive E. coli isolates were resistant to colistin (7).
Additionally, Ramezani et al. studied the antibiotic resistance of E. coli isolates from stool samples through experimental and molecular analyses (23). They reported that most E. coli isolates were resistant to erythromycin (100%) and ampicillin (94%) but were sensitive to nitrofurantoin (96%) and amikacin (100%). The E-test results showed the greatest resistance to erythromycin (100%), and the highest sensitivity was observed to gentamicin and amikacin (100%) (23).
5.1. Conclusions
Due to the increasing resistance to certain antibiotics and the wide range of changes in drug effectiveness, drug susceptibility testing has become crucial to prevent the escalation of drug-resistant infections. The high prevalence of ECAR and the high percentage of antibiotic resistance genes in clinical and livestock samples in the present study may indicate the overuse of antibiotics. However, this study has some limitations, including a small sample size, financial constraints, and the exclusion of other antibiotics. Future studies could explore other genes involved in antibiotic resistance to provide a more comprehensive understanding and suggest suitable solutions.