1. Background
Carbapenem-resistant Enterobacteriaceae (CRE) are categorized as critical priority pathogens by the World Health Organization (WHO) and the Centers for Disease Control and Prevention (CDC) due to their high resistance to last-resort antibiotics and their role in hospital-acquired infections (1, 2). The key mechanism behind this resistance is the production of carbapenemase enzymes, such as Klebsiella pneumoniae carbapenemase (KPC), New Delhi Metallo-β-lactamase (NDM), Verona Integron-Mediated Metallo-β-lactamase (VIM), Imipenem (IMP), and Oxacillinase (OXA), which hydrolyze carbapenems and render them ineffective (3, 4).
Healthcare-associated infections (HAIs) can arise from both direct clinical interventions and exposure to contaminated hospital environments (5). Over the past two decades, more than 30 studies have reported carbapenem-resistant organisms in hospital water systems, particularly in faucets, sinks, and drains (6). Sinks have emerged as critical reservoirs for multidrug-resistant organisms (MDROs), including Carbapenemase-Producing Enterobacteriaceae (CPE), Carbapenem-Resistant Acinetobacter baumannii (CRAB), and Pseudomonas aeruginosa (CRPA), all of which pose a transmission risk to patients (6, 7).
Sink drains are increasingly recognized as environmental niches for persistent colonization by resistant bacteria (6-8). Transmission from sinks to patients may occur via splash-back or aerosolized droplets during faucet use, which can contaminate nearby surfaces and equipment (8). A study by Kotay et al. demonstrated that droplet-mediated dispersion is a major transmission route from contaminated sink traps (8). Moreover, documented outbreaks have shown recontamination of sinks even after replacement, suggesting biofilm persistence in drainage systems (9).
Carbapenemase genes are frequently located on mobile genetic elements such as plasmids, facilitating their spread between bacteria and across healthcare environments (6, 10). Molecular methods like PCR have proven effective in detecting these genes with high sensitivity and specificity, despite their cost and technical demands (11). In Indonesia, environmental surveillance of carbapenemase in hospital settings remains limited. This study addresses that gap by investigating the presence and distribution of carbapenemase genes in sinks from various wards in general hospitals in Makassar.
2. Objectives
The aim of this study was to determine the presence of carbapenemase in sinks in the emergency room (ER), intensive care unit (ICU), and inpatient rooms using molecular methods in general hospitals in Makassar city.
3. Methods
This cross-sectional observational study analyzed 63 sink specimens collected from 19 general hospitals in Makassar City, Indonesia. Samples were processed at Hasanuddin University Hospital’s Microbiology Laboratory. Eligible sinks were functional and located in ERs, ICUs, or inpatient wards. Non-functional sinks or those without running water were excluded. Ethical approval was obtained from Hasanuddin University’s Faculty of Medicine IRB (Approval No. UH24050398).
3.1. Sink Sampling
From 19 hospitals, at least three sinks were sampled — one each from the ER, ICU, and inpatient ward. Sink drains were swabbed using COPAN Flocked Eswabs per manufacturer instructions. Samples were transported in Amies medium on ice and cultured on MacConkey agar (12).
3.2. Laboratory Methods
Swabs were processed within 24 hours by streaking on MacConkey agar and incubated overnight at 37°C. Colonies were examined for lactose fermentation. PCR was performed on grown colonies to identify gram-negative bacteria (Escherichia coli, Enterobacter sp., Klebsiella sp., K. pneumoniae, P. aeruginosa) and detect carbapenemase genes (blaIMP, blaVIM, blaNDM, blaOXA, blaKPC).
DNA was extracted using the gSYNC™ kit. PCR mixtures contained GoTaq Master Mix, primer sets specific for carbapenemase genes or bacterial identification, nuclease-free water, and DNA template, with a final volume of 50 µL. Amplification used a BioRad Thermal Cycler with the following protocol: Initial denaturation at 94°C for 2 minutes; 30 cycles of 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 90 seconds; and a final extension at 72°C for 10 minutes. PCR products were analyzed by electrophoresis on 2% agarose gels stained with ethidium bromide at 100 V for 30 minutes (12-15).
3.3. Data Collection
Swab samples from sinks were collected from all hospitals between July and August 2024. Staff knowledgeable about hospital characteristics were interviewed, and photographs of sampled sinks were taken.
3.4. Data Analysis
Descriptive analysis was used to characterize the sinks in the hospitals in the study. Statistical analysis was conducted using Microsoft Excel.
4. Results
This study involved 19 general hospitals out of 25 general hospitals in Makassar city. Most of them were class B public hospitals, namely 11 hospitals (57.9%). A total of 7 (36.8%) were class C hospitals, and 1 (5.3%) was a class A hospital.
4.1. Sink Characteristics
Most of the sink swab samples were taken from inpatient rooms, totaling 24 locations. About 90% of the sinks were clean and equipped with hand soap facilities. Additional facilities such as paper towels were available at 28 sinks (44.44%). The main function of the sinks in this study was to wash the hands of health workers. A total of 21 sinks (33.33%) were located close to the patient's bed, mostly with a distance of more than 2 meters and equipped with a dividing curtain (Figure 1). The majority of the sinks, 90%, were made of ceramic material. Table 1 provides more information about the characteristics of the sinks that were the subject of this study.
| Sink Characteristics | Amount (N = 63) | % |
|---|---|---|
| Sink location | ||
| Emergency room | 20 | 31.7 |
| Intensive care unit | 19 | 30.2 |
| Surgical ward | 24 | 38.1 |
| Cleanliness of the sink | ||
| Sink appears clean | 57 | 90.48 |
| Sink does not appear clean | 6 | 9.52 |
| Availability of hand soap | ||
| Soap cleanser available at sink | 59 | 93.65 |
| Soap cleanse unavailable | 4 | 6.35 |
| Availability of tissues | ||
| Tissues available at sink | 28 | 44.44 |
| Tissues unavailable | 35 | 55.56 |
| Sink function | ||
| Only staff handwashing | 47 | 74.6 |
| Handwashing staff and patient | 16 | 25.4 |
| Washing medical devices | 15 | 23.81 |
| Presence of bed near the sink | ||
| There is a bed near the sink | 21 | 33.33 |
| No bed is near the sink | 42 | 66.67 |
| Presence of curtain/bed divider with sink | ||
| There is a curtain between the bed and the sink | 14 | 66.67 |
| No curtain between the bed and sink | 7 | 33.33 |
| Distance between sink and patient bed (m) | ||
| < 2 | 6 | 28.57 |
| > 2 | 15 | 71.43 |
| Sink material | ||
| Ceramics | 57 | 90.48 |
| Stainless steel | 6 | 9.52 |
4.2. Hospitals Contaminated with Gram-Negative Bacteria and Carbapenemase Genes in Sinks
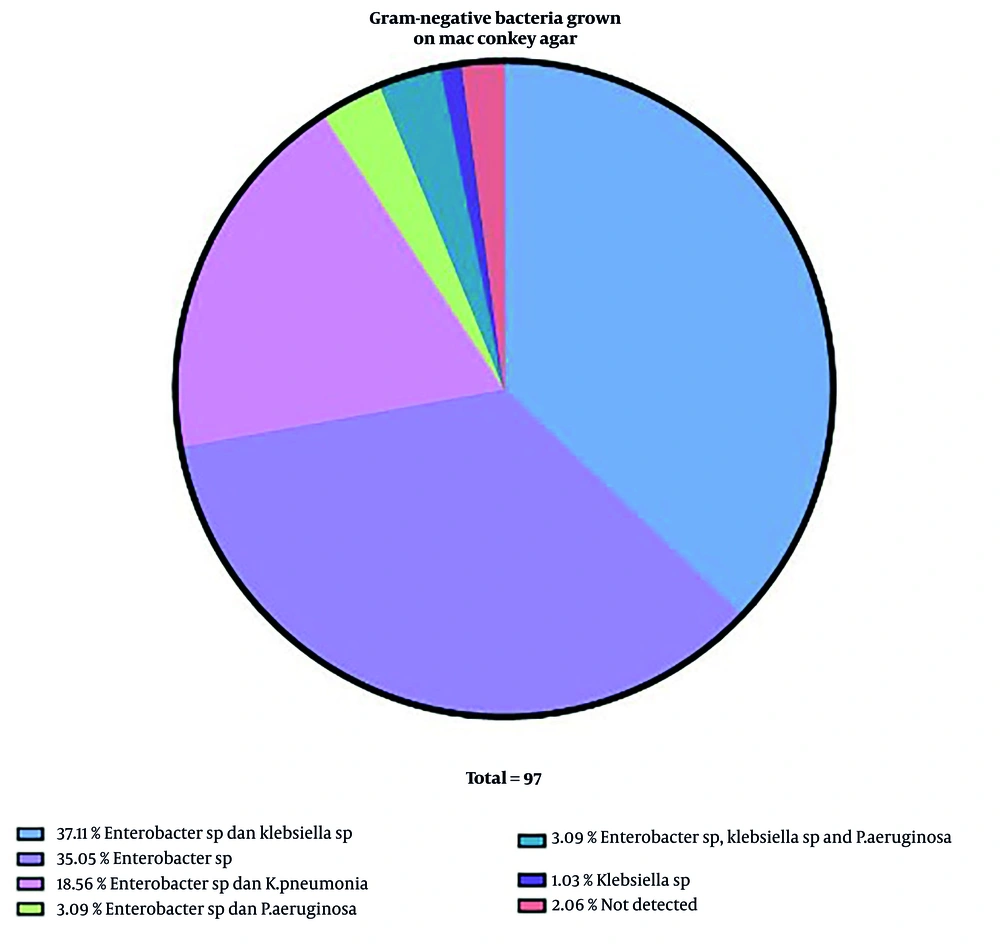
This study showed that of the 19 public hospitals studied, 10 hospitals (52.6%) were found to have carbapenemase genes in sink swab samples. This figure presents the distribution of gram-negative bacterial isolates cultured on MacConkey agar. The most prevalent combinations were Enterobacter sp. and Klebsiella sp. (37.11%) and Enterobacter sp. alone (35.05%). Other notable combinations included Enterobacter sp. with K. pneumoniae (18.56%) and Enterobacter sp. with P. aeruginosa (3.09%). A small percentage of isolates were solely Klebsiella sp. (1.03%) or yielded no detectable bacteria (2.06%). The total number of isolates analyzed was 97. These results highlight the dominance of Enterobacter and Klebsiella species in the sampled environments, which are common opportunistic pathogens in healthcare settings (Figure 2).
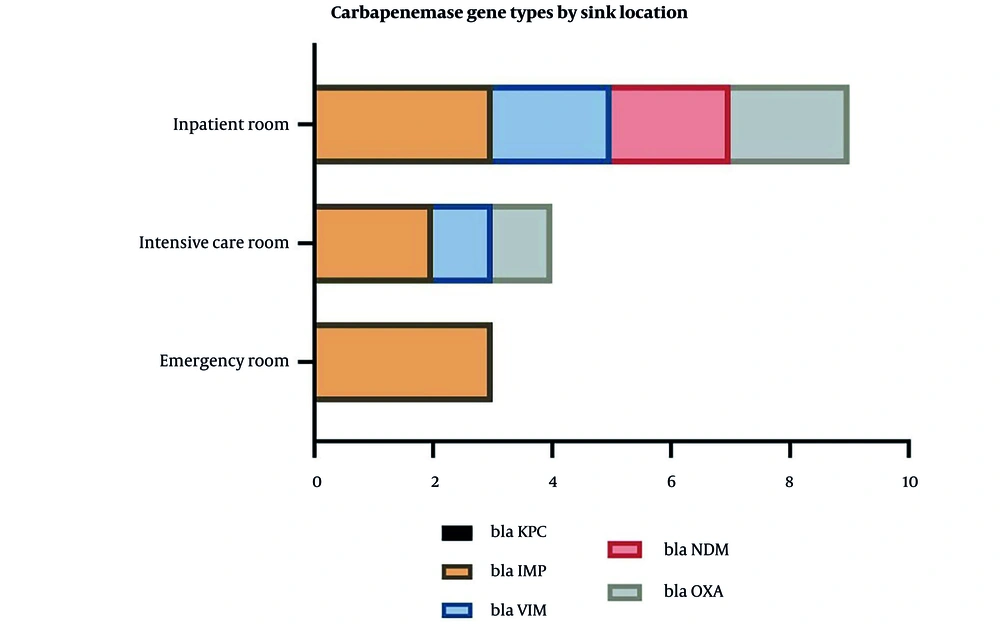
This figure illustrates the distribution of carbapenemase gene types across different hospital sink locations: Inpatient rooms, intensive care rooms, and ERs. The most frequently detected gene was blaKPC, followed by blaIMP, blaVIM, blaNDM, and blaOXA. The data suggests variability in gene prevalence by location, with inpatient and intensive care rooms showing higher detection rates for certain genes (e.g., blaKPC). This underscores the potential role of hospital sinks as reservoirs for carbapenem-resistant genes, particularly in high-risk areas like ICUs (Figure 3).
4.3. Distribution Pattern of Carbapenemase Genes in Sinks
The results showed that the distribution of carbapenemase genes in sinks in general hospitals in Makassar city was spread across 8 different sub-districts. The carbapenemase gene contamination of these hospital sinks showed a varied distribution pattern, indicating variations in the types of resistant bacteria that may be related to the conditions and infection prevention practices in each region.
The VIM gene was found in sinks in hospitals located in Tamalanrea, Ujung Pandang, and Manggala sub-districts, indicating the potential spread of highly resistant bacteria in these sub-districts. The IMP-resistant Pseudomonas carbapenemase gene was detected in hospital sinks located in four sub-districts, namely Biringkanaya, Tamalanrea, Ujung Pandang, and Manggala, which may indicate the presence of more widespread contamination in different parts of the city, especially for this imipenem-resistant gene.
In addition, the OXA gene, which also contributes to carbapenem antibiotic resistance, was found in hospitals located in Ujung Tanah, Rappocini, and Tamalate sub-districts, suggesting the spread of OXA gene contamination in these areas. The NDM gene, which is notorious for its high resistance and global spread, was found in the sinks of hospitals located in Wajo and Tamalanrea sub-districts.
4.4. Proportion of Contaminated Sinks by Location and Types of Carbapenemase in Hospital Sinks
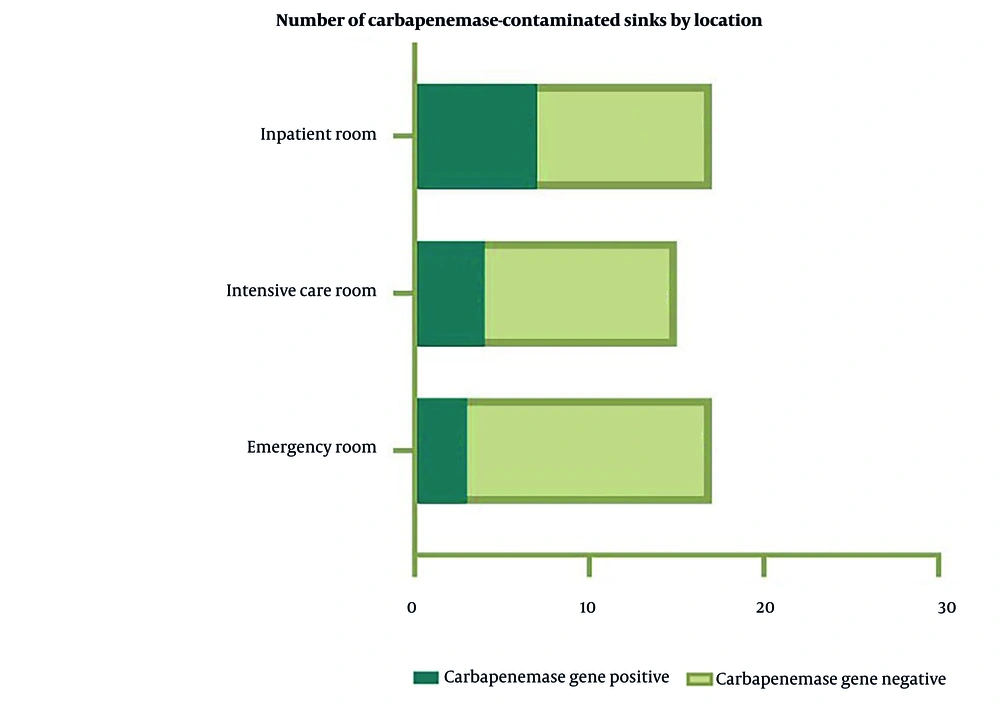
The figure compares the number of sinks testing positive or negative for carbapenemase genes across inpatient rooms, intensive care rooms, and ERs. Inpatient rooms had 7 positive and 17 negative sinks, intensive care rooms had 4 positive and 15 negative sinks, and ERs had 3 positive and 17 negative sinks. While the majority of sinks were negative, the presence of positive sinks in all locations — especially inpatient and intensive care rooms — indicates persistent contamination risks. The high number of negative sinks suggests potential success in infection control measures, but the positive cases warrant further attention (Figure 4).
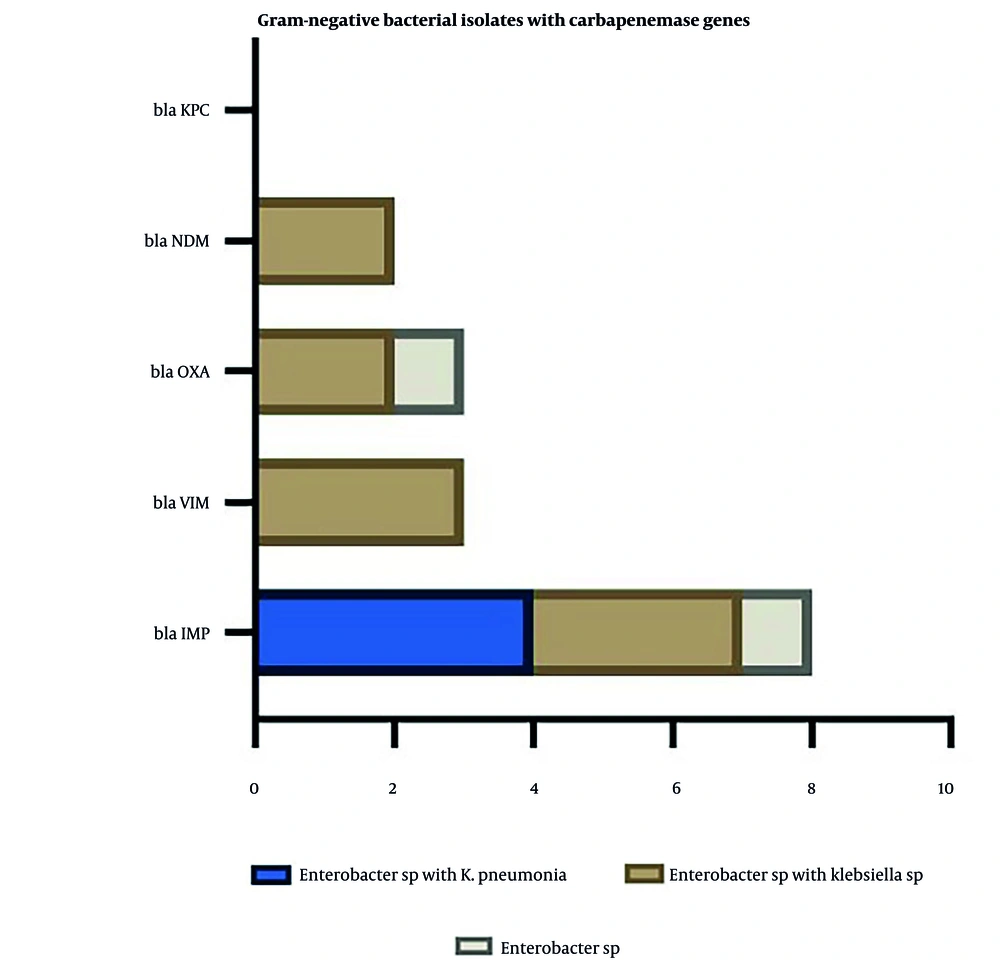
This figure correlates specific gram-negative bacterial isolates with detected carbapenemase genes. The gene blaKPC was the most common, followed by blaNDM, blaOXA, blaVIM, and blaIMP. The isolates most frequently associated with these genes were Enterobacter sp. with K. pneumoniae, Enterobacter sp. with Klebsiella sp., and Enterobacter sp. alone. This suggests that Enterobacter and Klebsiella species are key carriers of carbapenemase genes, which could contribute to the spread of antibiotic resistance in clinical environments (Figure 5).
5. Discussion
The increasing use of antibiotics has led to an increase in the number of antibiotic-resistant bacteria, such as carbapenemase-producing gram-negative bacteria. In Indonesia, research on carbapenemase-producing bacteria in the hospital environment is still very limited. Meanwhile, the hospital environment can be a source of resistant bacteria (16). Our findings show that 10 out of 19 general hospitals (52.6%) harbored carbapenemase genes in sink swab samples, distributed across 8 sub-districts. This underscores a significant environmental reservoir of resistance within hospitals, particularly concerning given the critical role of carbapenems in managing severe infections. The widespread distribution of genes reflects a diverse resistance pattern that likely corresponds with local antibiotic usage and infection control practices (9).
The highest contamination rates were observed in sinks located in inpatient rooms (29%), followed by ICUs (21%) and emergency departments (15%). Inpatient rooms typically experience more prolonged stays, higher foot traffic, and possibly less stringent environmental cleaning protocols compared to ICUs. These factors increase the risk of biofilm development and cross-contamination, as also reported in prior studies (17).
Pathogenic bacteria are often present in sinks in healthcare settings, including bacteria that have genes for resistance to multiple antibiotics. Sinks have been identified as a source of hospital-to-patient transmission and infection outbreaks, especially sinks located less than 1 meter from the patient, which can serve as an important reservoir for multidrug-resistant pathogens such as Carbapenem Resistant Organisms (CRO) (18). The physical proximity of patient beds to sinks was another noteworthy risk factor. In 21 hospitals, beds were located near sinks, with at least one hospital having a distance of less than 2 meters — conditions that facilitate bacterial transmission via splash or aerosol. Previous studies in Belgium and South Africa support this mechanism, where carbapenemase genes such as blaNDM and blaVIM were detected in sink aerosols despite no direct link to colonized patients (19, 20). Thus, sinks not only act as passive reservoirs but may actively contribute to HAIs through environmental contamination.
Supporting this, the study identified a variety of carbapenem-resistant species, including P. aeruginosa, K. pneumoniae, Enterobacter spp., and A. baumannii. These findings are consistent with previous multicenter studies in Ohio and Asia that reported frequent colonization of sinks with CPE and non-fermenters (6, 21-23). The presence of such organisms in moist environments such as P-traps and drains can lead to persistent colonization and periodic release of contaminated aerosols, especially during water use.
Notably, blaIMP was the most prevalent gene identified, found predominantly in ICU and inpatient areas. Other Metallo-β-lactamase genes (blaVIM, blaNDM) and the Ambler class D gene blaOXA were also detected. Interestingly, the class A gene blaKPC was not found in any sample, which contrasts with findings from Israel and parts of the US, where blaKPC is more dominant (9, 24, 25). This suggests regional differences in resistance gene epidemiology, possibly driven by distinct antimicrobial stewardship and infection control protocols.
Our findings align with surveillance data from Southeast Asia, where blaNDM, blaIMP, and blaVIM are the most commonly reported carbapenemase genes (25). For instance, a Philippine study from 2013 - 2016 identified blaNDM in 33% of carbapenem-resistant isolates. In India, blaNDM and blaOXA-23 dominate among Enterobacteriaceae and A. baumannii, respectively.
The study conducted on sink swab samples from general hospitals in Makassar City identified significant carbapenemase contamination, predominantly in hospitalization areas. Carbapenemase genes detected were categorized under Ambler class B (Metallo-β-lactamases: IMP, VIM, NDM) and class D (OXA), with the IMP gene being the most prevalent. The absence of the Ambler class A KPC gene was notable.
5.1. Conclusions
This study demonstrates that more than half of the general hospitals in Makassar have sinks contaminated with carbapenemase genes, particularly the IMP gene, with the highest contamination occurring in inpatient areas. The most common bacteria found were Enterobacter sp. and Klebsiella sp., which harbored diverse resistance genes. These findings underscore the role of hospital sinks as environmental reservoirs for antibiotic-resistant bacteria and highlight the urgent need for robust infection control and sink maintenance strategies. Strengthening surveillance and applying strict disinfection protocols are essential to prevent nosocomial transmission of carbapenem-resistant organisms.
5.2. Limitations
This study has several limitations. First, the relatively small sample size may limit the generalizability of the findings to all hospitals in Indonesia. Secondly, not all factors that may affect contamination (such as cleaning schedules, sink materials, and usage practices) could be controlled or collected equally. In addition, the association between the presence of resistance genes and hospital factors was not statistically analyzed due to the descriptive study design.
Continuous surveillance of the epidemiological dynamics of carbapenemase genes in sinks is critical to understanding their dissemination within our hospital. Enhanced focus on strengthening nosocomial infection prevention and control strategies is imperative. Furthermore, the investigation of additional carbapenemase-producing genes is warranted to comprehensively address the potential threats posed by antimicrobial resistance.