1. Background
Hepatitis B virus (HBV) has infected more than 2 billion people in the world (1), and it has been estimated that 350 million people will progress to the chronic phase of the disease (2). This partially double stranded DNA virus uses Reverse Transcriptase enzyme for replication. Reverse Transcriptase is susceptible to error in function and is incapable of proof reading after error in replication. Therefore mutation rate in HBV genome is 10-fold higher than other DNA viruses which explains the existence of different strains of this virus (3). Hepatitis B Viral genome consists of four overlapping open reading frames (ORF) (4). Hence, a mutation in each of the hepatitis B genes may lead to changes in other genes.
A to H genotypes have been described based on a greater than 8% difference among HBV strains (5). On the other hand, based on sequence variations in hepatitis B surface antigen (HBsAg), nine subtypes of HBV, namely ayw1, ayw2, ayw3, ayw4, ayr, adw2, adw4q-, adrq+, and adrq- have been explained. Genotype D and subtype ayw2 are the most prevalent strains which have been reported from the Middle East (6). Moreover, in the Middle East a range of endemicity for hepatitis B has been reported from different countries. For example Pakistan and Iran (7) have low endemicity (8), Egypt (9) and Turkey (10) have intermediate endemicity and Yemen (11) and Oman (12) have high endemicity of hepatitis B infection.
The most common method for HBV diagnosis is the ELISA technique for HBsAg (13). Some mutations in HBsAg genome can lead to incomplete HBsAg production (14). One of these mutations, premature stop codon mutation, helps the virus get rid of its immunogenic part of HBsAg (15, 16) and offers the opportunity to escape the host immune system. This mutation in the S gene is replacement of Thimidine (T) to adenosine (A) at position 207 that changes codon TGT to TGA, a stop codon that cuts the amino acid formation at position 69 of HBs Ag (17). Therefore, the occurrence of this mutation in a virus may render it undetectable by the ELISA methods for HBsAg detection. Due to overlapping of polymerase gene with the S gene, the amino-acid of Serine is replaced with Threonine at position 413 of reverse transcriptase gene (17).
2. Objectives
Premature stop codon mutation has been previously reported from the Middle East (18). As blood products are screened by the ELISA methods for HBsAg (19), the frequency of premature stop codon mutation in blood donors may cause serious concerns about the safety of the screening protocol (17, 20). Therefore, we performed a thorough search in the Genbank database on the submitted HBV genome from Middle Eastern countries.
3. Materials and Methods
In this study, we used the PubMed central (National center for biotechnology information) web site, Genbank subsection, for collecting genomic data using the following key words: hepatitis B, S gene and the name of each Middle Eastern countries. According to MESH section of NCBI, Middle Eastern countries include Afghanistan, Bahrein, Iran, Iraq, Israel, Jordan, Kuwait, Lebanon, Oman, Qatar, Saudi Arabia, Syria, Turkey, United Arab Emirates and Yemen. The search was performed on the 1th of March 2012. Bioedit software version 7.1.3 was used to evaluate the occurrence of premature stop codon in S gene at any position. Hepatitis B Virus genotype and sub genotype was recorded for each submitted sequence. HBV subtype was distinguished based on specific sequences of deduced amino acids of each sequence.
4. Results
4.1. Hepatitis B Virus Sequences Reported From Middle Eastern Countries
Pubmed search retrieved 1341 HBV S gene sequences isolated from Middle East countries (Afghanistan, Bahrain, Iran, Iraq, Israel, Jordan, Kuwait, Lebanon, Oman, Qatar, Saudi Arabia, Syria, Turkey, United Arab Emirates and Yemen). From 1341 HBV S gene sequences, 12 sequences were submitted from Afghanistan, 46 sequences from Sudan, 320 sequences from Turkey, 711 sequences from Iran, 61 sequences from Egypt, 88 sequences from the UAE, 64 sequences from Morocco and 33 sequences from Yemen. Only three HBV genomes were reported from Israel, two from Iraq and one from Syria .We could not find any HBV S gene sequences from Bahrain, Qatar, Oman, Kuwait, Lebanon, Jordan, Libya and Saudi Arabia. All sequences from Afghanistan were reported by Iranian research centers. None of the UAE sequences qualified for the inclusion criteria and hence were omitted from the current study. Therefore, 1253 out of 1341 sequences were evaluated in this study.
4.2. HBsAg Positivity Among Middle Eastern HBV Infected Patients
The information about HBsAg statues of patients, who had their HBV sequences submitted to Genbank, was not accessible for all candidates. Based on articles, which have reported on these sequences, 70% of them were HBsAg positive and others have no documents about their HBsAg condition.
4.3. Pre Mature Stop Codon Mutation Among HBV Derived From Different Countries
We observed premature stop codon in 5.7% of Turkish sequences, 3.8% of Iranian sequences, 3% of sequences from Yemen and 50% of sequences from Iraq (Table 1).
| Country | Populationa | HBVbS Gene, No. | PSCb,c, No. (%) | PSC in Blood Donors, No. |
|---|---|---|---|---|
| Afghanistan | 34,385,068 | 12 | NDRb | 0 |
| Sudan | 43,552,000 | 46 | 0 (0) | 0 |
| Turkey | 73,722,988 | 320 | 4 (6) | 0 |
| Iran | 75,416,000 | 711 | 17 (4) | 1 |
| Egypt | 80,438,000 | 61 | 0 (0) | 0 |
| Bahrain | 807,000 | 0 | NAb | NA |
| Qatar | 1,696,563 | 0 | NA | NA |
| Oman | 905,000 | 0 | NA | NA |
| Kuwait | 3,051,000 | 0 | NA | NA |
| Lebanon | 4,255,000 | 0 | NA | NA |
| UAEb | 4,707,000 | 88 | Not qualified | 0 |
| Jordan | 6,187,000 | 0 | NA | NA |
| Libya | 6,355,000 | 0 | NA | NA |
| Morocco | 32,000,200 | 64 | 0 (0) | 0 |
| Yemen | 24,256,000 | 33 | 1 (3) | 1 |
| Israel | 7,797,400 | 3 | 0 (0) | 0 |
| Iraq | 31,467,000 | 2 | 1 (50) | 0 |
| Syria | 22,505,000 | 1 | 0 (0) | 0 |
| KSAb | 27,136,977 | 0 | NA | NA |
Frequency of HBV S Gene and Premature Stop Codon in Middle-Eastern Countries
As reported in Genbank, in some cases information on the source of the virus was not available. This was observed in one S gene mutations from Turkey and two HBV sequences from Iraq, where the latter included one premature stop codon mutation.
4.4. Pre Mature Stop Codon Mutation Among HBV Derived From Blood Donors
Among HBV S mutant genes which were submitted from Yemen, one sequence was derived from a blood donor. In Iran, out of 17 mutant sequences, one strain was derived from a blood donor (Table 1).
4.5. HBV Genotype and Subtype of S Gene Pre-Mature Stop Codon Mutants
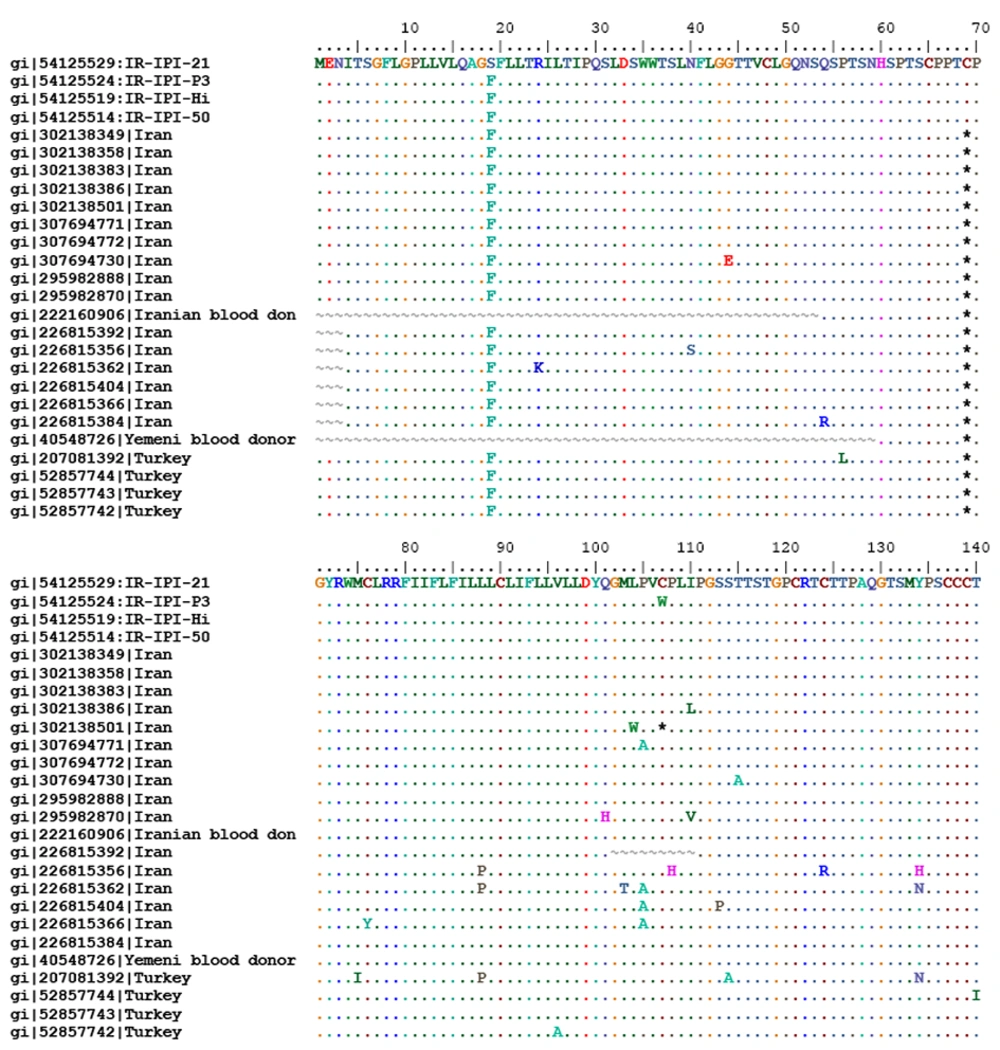
All HBV sequences with premature stop codon were genotype D and based on Arg122 and Lys160 and Pro127 in the deduced amino acid sequences of the S gene were categorized as subtype ayw2 (Figure 1).
5. Discussion
Despite effective vaccination, hepatitis B virus is a universal health problem (21). Hepatitis B new infections and occult hepatitis B (OBI) are still reported from all around the world (22-25). Nowadays, ELISA techniques are the most frequent methods for primary diagnosis of HBV infection in suspected patients and among blood donors (26). Therefore, better understanding of ELISA misdiagnosis for HBV infection helps us to more accurately diagnose infected patients and control new infections (27). In the Middle Eastern countries, HBV transmission mostly happens inside families due to socio-economic statues and population distribution (28). This means that infected patients usually live with the HBV virus for their whole life (29) and this provides an opportunity for the virus to produce strains with an ability to escape from the host immune system (25). One of these types of mutations is S gene premature stop codon. Since ELISA HBsAg kits are designed to identify wild type of HBsAg, mutant type of this protein are antigenically modified and are undetectable by this technique (26). Some researchers have shown that variant surface antigens could not be detected by monoclonal antibodies of surface antigen (16) while others have suggested that the most sensitive HBsAg assays are not helpful for detection (30). In this study we realized that 70% of patients with this type of mutation have HBsAg. Since in Genbank there was no information about other HBsAg statues, we are not able to judge the HBsAg kit and its reliability for mutant HBsAg diagnosis. However, from available reports (14, 17) we can gather that this mutation can produce truncated HBs protein and so is undetectable by available HBsAg kits.
As depicted in Table 1, there are unfortunately only a few data and submitted sequences from many countries with considerable population. No HBV S gene sequences were submitted from Saudi Arabia, Libya, Jordan, Lebanon, Bahrain, Qatar, Oman and Kuwait that did not allow the opportunity to determine the prevalence of premature stop codon in these countries. Most of the HBV sequences were reported from Iran (31-33) and Turkey (34, 35). Only a few sequences were reported from Syria, Israel and Iraq, resulting in a wide confidence interval in their estimation of S gene mutation rate. Premature stop codon mutation was observed among most Middle Eastern countries that had submitted evaluable HBV genomes to Genbank. This mutation was previously described by Vaezjalali et al. (18) for Iranian HBV patients with cirrhosis. The interesting point is that although we observed this mutation in submitted HBV genomes from Iran, Iraq, Yemen and Turkey, yet we could not find any comment about their existence in the published literature from the respective countries. In other words, researchers seem to have overlooked the importance of this mutation.
In blast search of the sequences, we observed premature stop codon mutation in blood donors from Yemen (accession number: AY486410) and Iran (accession number: 222160906). The existence of these mutations in healthy blood donors, previously reported by Datta et al. (17), may be an alarming sign for the possibility of HBV transmission through transfusion if HBsAg ELISA techniques are solely used for the screening of blood products. There are concerns about the ability of the mutant virus to obtain appropriate surface proteins to exit from infected cells and be able to be transmitted to other individuals (36). The existence of wild type viruses in patients with mutant strains enables the virus to exit the infected cells and so these patients are infective (14). In this study we were not able to identify the source of isolated HBV S gene due to inadequacy of information recorded in Genbank. Complete information about the submitted sequences will be useful, to gain a better access to data of HBV genomes for future analysis. In conclusion, we observed premature stop codon S gene mutations among HBV genomes from most Middle Eastern countries, which were not commented on before. In contrast to other studies, it seems that the premature stop codon 69 may not alter HBsAg-positivity by ELISA. More studies on antigenicity of the mutant and wild type form of HBsAg can be helpful to solve this paradox.