1. Background
Helicobacter pylori, a Gram-negative bacterium found on the luminal surface of the gastric epithelium, was first isolated by Warren and Marshall in 1983. It induces chronic inflammation of the underlying mucosa, chronic, acute, and atrophic gastritis and gastric, and duodenal ulcer diseases (1, 2). Treatment of the infecting strain and its eradication can lead to treatment of the diseases. Unsuccessful treatment of the bacterium and carriage of H. pylori is strongly associated with the risk of atrophic gastritis development, which is a precursor lesion to gastric cancer (3, 4). It is estimated that H. pylori-positive patients have a 10-20% lifetime risk of developing ulcer disease and a 1-2% risk of developing distal gastric cancer (5). Eradication of H. pylori is still a challenge, because of the rapidly increasing prevalence of multidrug resistant strains worldwide (6). In consequence of the increasing resistance of H. pylori against common antibiotics, next generation drugs, such as quinolones, will be required for eradication therapy in the future. Resistance of H. pylori to fluoroquinolones, happening through the effect of DNA gyrase A subunit on the basis of point mutations in the quinolone resistance-determining region of the gyrA gene, has recently become common (7, 8). In fact, H. pylori strains with reduced susceptibility to fluoroquinolones have a mutation at codon 87, Asn of the gyrA gene (9). New researches by Wang et al. in China, Chung et al. in Korea, and Chisholm et al. in UK, reported frequencies of 36.66 %, 48.27 % and 23.5 % for mutation of Asn87Lys (AAT > AAA and AAC > AAG) in all fluoroquinolone-resistant isolates, respectively. These results showed that frequency of these mutations was high in H. pylori isolates of different populations (10-12).
Usage of unprescribed drugs, inappropriate administration of quinolones for common infections by other bacteria, and spontaneous mutations happening during the bacterial infection cycle in the human body, can lead to emergence of newly developed resistant strains, which could be challenging (13). Rapid detection of these resistant strains in clinical samples is important for controlling their dissemination in the community. Due to the difficulty of the culture-based method for common antimicrobial susceptibility testing approach, development of rapid and simple molecular techniques can overcome this need. Because of the heterogeneity of gyrA gene among H. pylori strains in different geographic areas, we developed a rapid test based on an allele-specific PCR (AS-PCR) to detect more frequent mutations of Asn87Lys (AAT > AAA and AAC > AAG) in the gyrA gene among different H. pylori isolates in Iran. The gyrA mutations of H. pylori causing reduced susceptibility to fluoroquinolones could be detected by this method. The AS-PCR method for detection of the gyrA mutations in H. pylori can be useful for easy identification of the targeted fluoroquinolone-resistant strains of H. pylori in the clinical samples (3).
2. Objectives
The aim of this study was designing a simple and rapid AS-PCR-based method for detection of more frequent gyrA mutations at the Asn87Lys codon, responsible for emergence of fluoroquinolone resistance in H. pylori strains.
3. Patients and Methods
3.1. Bacterial Strains
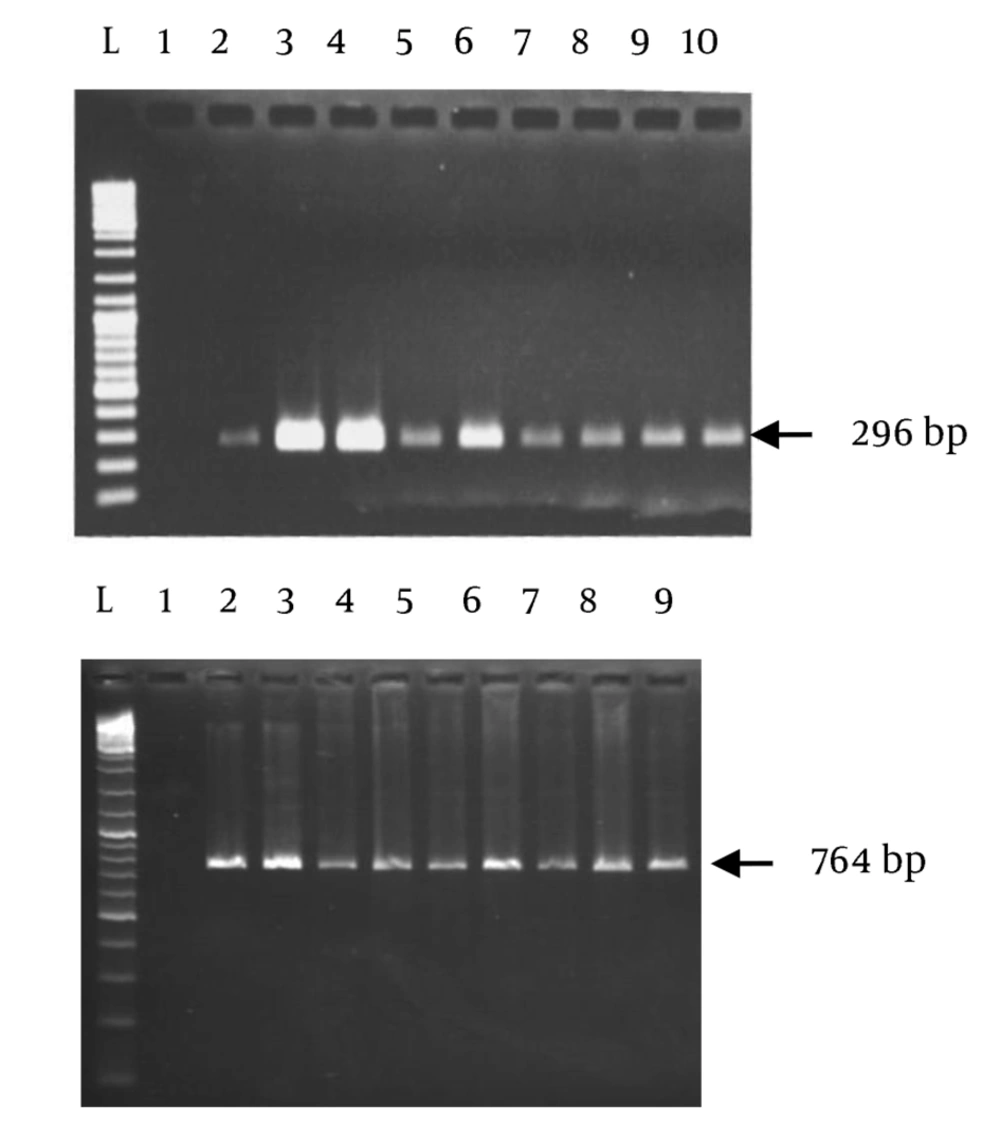
All of the used bacterial strains in this study were obtained from clinical biopsy samples in our laboratory at Gastroenterology and Liver Diseases Research Center, Tehran, Iran. The isolates were identified by culture and genus and species-specific PCR primers for 16s rRNA and glmM; the primer sequences are listed in Table 1 (14, 15). Susceptibility of the isolates to ciprofloxacin, as a member of fluoroquinolones, was determined based on the agar dilution method, as described before (16). PCR conditions were as follows: Total volume of PCR mixture was 25 mL; 0.12 pM of each primer, 2 mM/L MgCl2; 0.16 mM/L dNTP, 1.5 U Taq DNA polymerase, and 200 ng DNA sample. PCR was performed in a thermocycler (AG 22331; Eppendorf, Hamburg, Germany) under the following conditions: initial denaturation for 4 minutes at 94˚C followed by 30 cycles of 94˚C for 1 minute, 57˚C for 45 seconds, and 72˚C for 1 minute. After a final extension at 72˚C for 10 minutes, the PCR products were examined by electrophoresis on 1.2% agarose gels according to the standard procedures. Bacterial isolates were selected based on their previously reported resistance phenotype to ciprofloxacin and targeted mutations in gyrA gene.
| Primer Name | Primer Sequence |
|---|---|
| AAC > AAG | Forward: ACCACCCCCATGGCGATCCG |
| AAT > AAA | Forward: ACCACCCCCATGGCGATCCA |
| Reverse primer | Reverse: GTTAGGCAGACGGCTTGGTARAATA |
| glmM | Forward: GGATAAGCTTTTAGGGGTGTTAGGGG |
| Forward: GCTTACTTTCTAACACTAACGCGC | |
| 16S rDNA | Forward: GGCTATGACGGGTATCCGGC |
| Reverse: GCCGTGCAGCACCTGTTTTC |
3.2. Primer Design and Bioinformatic Investigations
Allele-specific primers were designed, in which the first nucleotide from the 3′ end match the site of the targeted point mutation. Furthermore, the second and third nucleotides from the 3′ end were designed to produce base pair mismatches, to attain high specificity in the AS-PCR for the mutant alleles. Specific binding capacity of the primers was checked by Iranian and other available H. pylori sequences in the GenBank database (JQ 323555-587) (16).
3.3. Site-Specific PCR Experiments
All H. pylori strains used in this study were confirmed by genus (16s rRNA) and species (glmM)-specific primers. DNA extraction of the isolates was performed by nucleospin tissue DNA extraction kit (MACHEREY-NAGEL GmbH & CO, KG). The control strains harboring mutations of Asn87Lys (AAT > AAA and AAC > AAG) were obtained from domestic resistance strains, previously sequenced in our laboratory. PCR conditions included denaturation at 95˚C for 4 minutes, followed by 25 cycles of denaturation at 94˚C for 1 minute, annealing at 51˚C for 45 seconds, and extension at 72˚C for 25 seconds, with final extension at 72˚C for 10 minutes for AAT > AAA primer, and denaturation at 95˚C for 4 minutes, followed by 25 cycles of denaturation at 94˚C for 1 minute, annealing at 53˚C for 45 seconds, and extension at 72˚C for 25 seconds, with final extension at 72˚C for 10 minutes for AAC > AAG Primer. PCR was performed in a thermocycler (AG 22331; Eppendorf, Hamburg, Germany). The PCR mixture (final volume of 25 μL) contained super Taq (Gen Fanavaran, Iran), 0.12 pM of each primer, 0.16 mM/L dNTP and 1.5 mM/L MgCl2. The primer sequences are listed in Table 1. Different clinical isolates with defined resistance phenotypes against fluoroquinolones were also analyzed for screening of the target mutations. The amplicons were analyzed by electrophoresis in 1% agarose gel (Fermentas, #R0491, Lithuania) in Tris-boric acid-EDTA buffer and stained with ethidium bromide. The gels were then photographed under the ultraviolet light (UVIdoc, UVItec Limited, Cambridge, UK).
3.4. Sequencing Experiments
To confirm the PCR results for each mutation, the amplicons were purified and subjected to sequencing by using the amplification primers and an applied biosystems (ABI) terminator cycle sequencing ready reaction kit (big Dye® terminator V3.1 cycle sequencing kit) on an ABI 3130xl genetic analyzer. The sequences obtained were edited manually and aligned using gene runner software (version 3.05). Analyses of the sequences were performed by Lasergene software (version 6.0), comparing with the reference sequences. The sequences were then submitted in GenBank.
4. Results
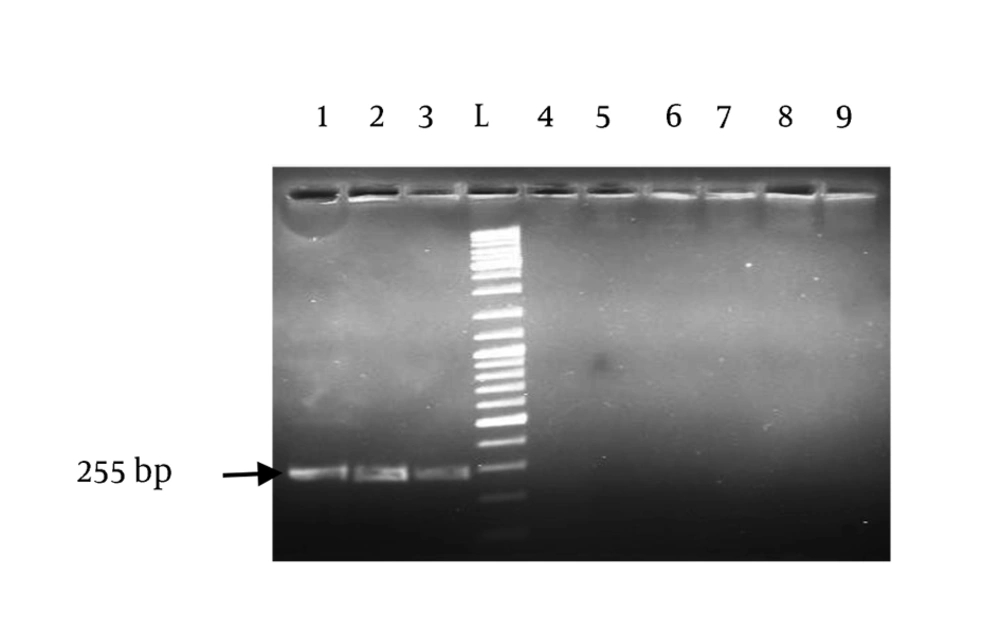
All the used strains in this study were from clinical samples. The isolates were identified as H. pylori by specific PCR for 16s rRNA and glmM (Figure 1). Presence of the targeted mutations had been confirmed in all the positive control strains (GenBank accession numbers: oc192:rcgldsh22_oc273:rcgldsh30_oc3:rcgldsh13). Bioinformatic analyses of the designed primers showed their efficacies with some criteria as presence of no bulge loop and internal loop, and optimal 3′ ∆G range. As shown in Figure 1, the primers clearly differentiated the wild type strains from the gyrA mutants. Unspecific bands were present in some strains. To delete these bands, concentration of MgCl2, extension time and number of cycles were changed in different PCR reactions. The best results were obtained at 1 mM MgCl2 concentration. Point mutations in the gyrA gene at codon Asn87Lys (AAT > AAA and AAC > AAG) were detected in all standard resistant strains and some clinical isolates with previously determined resistance phenotypes for fluoroquinolones. Similarity of the used annealing temperature and PCR conditions for these two primers, proposed their capacity for usage in same PCR reactions.
The products were amplified with allele-specific primers. Lane 1, positive control (OC3) for AAC > AAG mutation; lane 2 and lane 3, positive controls (OC192, OC273) for AAT > AAA mutation; lane 4 and lane 5, negative controls for AAC > AAG and AAT > AAA mutations, respectively; lanes 6, 7, 8, 9, randomly selected clinical isolates.
5. Discussion
Diverse assays have been developed to investigate genetic polymorphisms in human pathogens, including PCR-restriction fragment length polymorphism analysis, AS-PCR, multiplex PCR, single-strand confirmation polymorphism analysis, oligonucleotide ligation assay, and real-time PCR (9). AS-PCR is a simple, cost-effective and excellent genotyping method in this regard. For mutations in the gyrA gene of H. pylori, AS-PCR does not require restriction enzyme cleavage, purification of PCR products, or a real time PCR machine that can increase its application for routine tests in clinical laboratories. However, this method has some limitations, such as designation of primers at the boundary of the targeted mutations, which can affect the PCR results. In this study, PCR amplification was performed with allele-specific primers, in which the first nucleotide from the 3′ end was designed to match the site of the point mutation; furthermore, the second and third nucleotides from the 3′ end were designed to produce two mismatches to attain high specificity in the AS-PCR for the mutated alleles. The results showed high-quality, reproducibility and particularity along with the tested isolates. A number of the resistant isolates did not present positive PCR results for these mutations, proposing existence of other mutations in gyrA or other resistance mechanisms among the studied samples. This limitation suggested design of new primers as well as examination of other mutations. In a study by Nishizawa et al. heterogenicity of H. pylori strains in different geographic areas was presented as the main reason for weakness of AS-PCR in the cases of noted mutations in the gyrA gene (9, 17). The developed technique is useful for finding out the bacterial susceptibility to fluoroquinolones within several hours. Diverse mutation levels of H. pylori strains in different geographic regions may cause assay failure in detecting other genetic gyrA variants found in different countries (18). Designing allele-specific primers for other common mutations, responsible for development of resistance phenotypes in H. pylori strains against this group of antibiotics, helps us to improve their eradication by rapid detection in the laboratories.
In conclusion, according to the high frequency of gyrA mutations at the Asn87Lys codon among fluoroquinolone-resistant strains of H. pylori, we developed a reliable PCR-based technique to detect these mutations without bacterial culture and minimum inhibitory concentration determination. Results of this study successfully confirmed specificity of the designed primers in detection of the targeted mutations.