1. Background
Tuberculosis (TB), caused by bacteria (Mycobacterium tuberculosis), is one of the most important global public health issues and remains a major cause of death worldwide, particularly in Asia and Africa (1, 2). In 2013, according to the global tuberculosis report from WHO, approximately nine million individuals infected with TB and 1.5 million died from the disease (3). It is supposed that one-third of world population is infected with TB, however, 10% of infected cases would develop clinical disease during their lifetime (4). TB is a complex disease and it is not well identified why most infected persons never develop active TB during their lifetime. Chitin is a polysaccharide presents as a structural component in the exoskeleton of insects as well as cell walls of certain fungi and nematodes (5). Humans do not have chitin, but express active chitotriosidase (CHIT1), which hydrolyzes chitin.
Human chitotriosidase (CHIT1) is a member of the chitinase family, a group of glycoside hydrolases that cleave chitin. The human CHIT1 gene is localized on chromosome 1q31-q32.5. CHIT1 is present in normal plasma and mainly secreted by activated macrophages (6). Its activity has been proposed as a biochemical marker of macrophage accumulation and activity in several lysosomal lipid storage diseases (7-11).
CHIT1 is secreted by activated macrophages, so its activity has been proposed as a biochemical marker of macrophage accumulation (6-12).
CHIT1 has been shown to be expressed during differentiation and maturation of DCs, indicating that CHIT1 plays an important role in the DCs immunoresponse (13). CHIT1 is a component of the innate immunity, which may play a role in defense against chitin-containing pathogens (14). During the development of acute/chronic inflammatory disorders, the CHIT1 activity increases significantly (6).
It has been shown that chitins stimulate macrophages to make IL-12, IL-18 and TNFα (15) and act as an immunological adjuvant (16). Shibata et al. (17) have shown that chitins had Th-1 adjuvant effect in developing Th1 immunity against a mycobacterial antigen. This indicates that CHIT1 might modulate the plasma level of chitins and regulate immune responses. CHIT1 deficiency could increase circulating chitin levels, which may increase in immune reactivity toward tuberculosis.
A 24 bp duplication in exon 10 in the CHIT1 gene activates a cryptic 3ˊ splice site generating an abnormally spliced mRNA (18). The spliced mRNA encodes an enzymatically inactive protein that lacks an internal stretch of 29 amino acids. A mild and approximately absent CHIT1 activity has been found in heterozygous and homozygous mutant individuals for the duplication, respectively (18).
To the best of our knowledge, there is only one report regarding the association of 24 bp duplication of CHIT1 gene and PTB (19).
2. Objectives
The present study aimed to assess the impact of 24 bp duplication of CHIT1 on PTB risk in an Iranian population.
3. Patients and Methods
One hundred seventy-seven patients diagnosed with PTB and 163 healthy subjects were enrolled in this study. The cases were selected from PTB patients admitted to a university-affiliated hospital (Bou-Ali Hospital, Zahedan, referral center for TB). The diagnosis of PTB was based on clinical symptoms, radiological evidence and bacteriological investigations such as sputum acid fast bacillus (AFB) smear positivity, culture and response to antituberculosis chemotherapy (20-23). All control subjects were unrelated adults selected without recent sign, symptom or history of TB from the same geographical origin, as patients with PTB.
The local ethics committee of Zahedan University of Medical Sciences approved the project (No. 93-293, Aug 27, 2014). An informed consent was obtained from all participants. DNA was extracted from peripheral blood by salting-out method as described previously (24).
In this study, polymerase chain reaction (PCR) was used for detection of 24 bp duplication of CHIT1 gene. The forward and reverse primers were 5`-GAAGAGGTAGCCAGGCTCTGG-3` and 5`-CTGCCGTAGCGTCTGGATGAG-3`, respectively.
PCR was performed using commercial master mix. In each 0.20 mL PCR reaction tube, 1 μL of genomic DNA (~ 100 ng/mL), 1 μL of each primers and 10 μL of 2X Prime Taq Premix (Genet Bio, Korea) and 7 μL dd H2O were added.
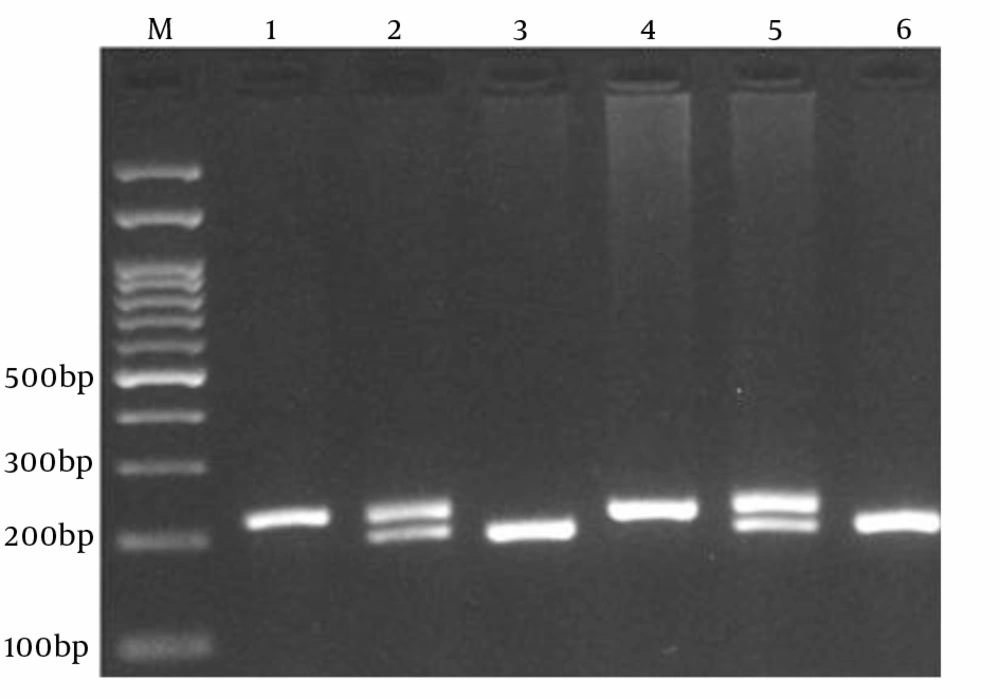
PCR cycling conditions were initial denaturation at 95˚C for 5 minutes followed by 30 cycles for 30 seconds at 95˚C, annealing at 68˚C for 30 seconds and extension at 72˚C for 30 seconds, with a final extension of 72˚C for 10 minutes. The PCR products were verified onto 2.5% Agarose gels containing 0.5 μg/mL ethidium bromide and observed under UV light (Figure 1). The genotype was determined by the presence or absence of a 195 bp wild-type (Wt) band or a 219 bp mutant (Mt) band. To certify genotyping quality, we regenotyped approximately 20% of the random samples and found no genotyping errors.
3.1. Statistical Analysis
Statistical analysis was performed using commercial software (SPSS for Windows, V 20, SPSS Inc., Chicago, IL, USA). Genotype and allelic frequencies were compared between the groups by chi-squared test. Logistic regression analysis was used to estimate odds ratio (OR) and 95% confidence intervals (CI) of genetic risk in PTB. A P value less than 0.05 was considered statistically significant.
4. Results
The study groups consisted of 173 patients with PTB (64 men and 109 women, aged 50.1 ± 20.6 years) and 164 healthy subjects (70 men and 94 women, aged 47.5 ± 15.3 years). There was no significant difference between the groups regarding sex and age (P = 0.317 and P = 0.191, respectively).
The genotype and allele frequencies of 24 bp duplication of CHIT1 gene are presented in Table 1.
| 24 Bp CHIT1 Duplication | Case | Control | OR, 95% CI | P Value |
|---|---|---|---|---|
| Wt/Wt | 74 (42.8) | 72 (43.9) | 1.00 | - |
| Wt/Mt | 78 (45.1) | 72 (43.9) | 1.05 (0.67 - 1.66) | 0.907 |
| Mt/Mt | 21 (12.1) | 20 (12.2) | 1.02 (0.51 - 2.04) | 0.925 |
| Alleles | ||||
| Wt | 226 (65.3) | 216 (65.8) | 1.00 | - |
| Mt | 120 (34.7) | 112 (34.2) | 1.02 (0.75 - 1.40) | 0.935 |
a The values are presented as No. (%).
Homozygous wild type, heterozygous and homozygous mutant frequencies of CHIT1 24 bp duplication polymorphism were 43.9%, 43.9% and 12.2% in controls and 42.8%, 45.1% and 12.1% in PTB patients, respectively. Chi-square comparison of PTB and control subjects and logistic regression analysis found no association between CHIT1 24 bp duplication genotype and PTB.
The mutant (Mt) allele frequency in controls and cases were 34.2% and 34.7%, respectively. The Mt allele was not associated with risk of PTB.
The CHIT1 variant in cases and controls were in HWE (χ2 = 0.004, P = 0.948 and χ2 = 0.929, P = 0.760, respectively)
5. Discussion
In the current study, we investigated the possible association between 24 bp duplication of CHIT1 gene and PTB in a sample of Iranian population in the southeast of Iran. Our findings showed no significant association between 24 bp duplication of CHIT1 gene and PTB.
Lee et al. (19) showed that 24 bp duplication in exon 10 of human CHIT1 gene leads to aberrant mRNA splicing and deletion of 87 nucleotides of the CHIT1 gene. They found that homozygotes 24 bp duplication of CHIT1 (deficient genotype) was significantly higher in patients with tuberculosis than control group in European ancestry, but not Asian ancestry. They found highest deficient allele frequency in Asian (0.56) and lower frequencies among European (0.17) and African ancestry (0.07). The mean activity of CHIT was nearly half normal in heterozygotes (wild/dup) and almost absent in subjects with homozygous mutant (dup/dup). The G354R and A442V polymorphisms of CHIT1 were significantly associated with reduced CHIT1 activity independent of 24 bp duplication.
CHIT-1 activity has been suggested as a biochemical marker of macrophage activation. It has been proposed that chitins stimulate macrophages to produce IL-12, IL-18 and TNF-α (15) and act as an immunological adjuvant as well as polyclonal B-lymphocyte activator (16). Chitins have been shown to have TH-1 adjuvant activity in immunity against mycobacterial antigen (17). Thus, CHIT activity could modulate the levels of chitins and regulate immune responses.
It has been proposed that CHIT1 is predominantly expressed in the human lung (25). Neutrophils and macrophages produce CHIT1 after toll-like receptor (TLR) stimulation (26). Macrophages also release CHIT1 after stimulation with interferon (IFN)-γ, tumor necrosis factor (TNF)-α or granulocyte-macrophage colony-stimulating factor (GM-CSF) (9).
Homozygous duplication of a 24 bp of CHIT1 gene abolishes enzyme activity. Heterozygous, homozygous and allele frequency of 24 bp duplication were 47.2%, 32.5% and 56.1% in Korean population, respectively (27). In the present study, we found the frequency of 43.9%, 12.2% and 34.2% for heterozygous, homozygous and allele of CHIT1 duplication. A significant association was found between 24 bp duplication in CHIT1 gene and asthma in North Indian population. The heterozygous (wild/dup) genotype as well as the combination of wild/dup and dup/dup increased the risk of asthma (28). The 24 bp duplication of CHIT1 gene was not associated with coronary artery disease (CAD) in Corsican population (29). It has been shown that CHIT could have a crucial role even in pathological conditions, such as coronary artery disease, acute ischemic stroke, cerebrovascular dementia and Alzheimer’s disease (30, 31).
Bargagli et al. (32) investigated serum CHIT1 activity in patients with sarcoidosis, PTB patients and healthy subjects. They found that CHIT1 activity was significantly higher in patients with sarcoidosis than controls or PTB patients. CHIT1 activity was not significantly different between PTB patients and controls.
In conclusion, our findings did not find an association between 24 bp duplication of CHIT1 gene and the risk of PTB in a sample of Iranian population. One limitation of this study was its relatively small sample size. Therefore, the results of this study should be interpreted with caution. Larger studies with different ethnicities are necessary to confirm our findings in various populations.